Figures & data
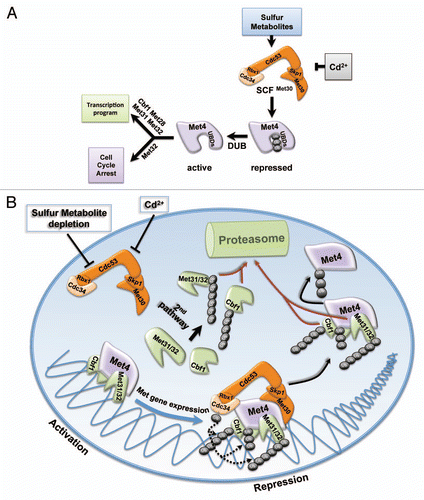
Figure 1 Regulation of sulfur metabolite homeostasis by ubiquitin-controlled transcription programs. (A) The ubiquitin ligase SCFMet30 links nutrient availability and heavy metal stress to cell cycle control and transcriptional response. The transcriptional activator Met4 is maintained in an inactive state by ubiquitylation. Ubiquitin-binding domains in the amino-terminal region of Met4 shield the ubiquitin chain from proteasome recognition and degradation. Nutrient or heavy metal stress block Met4 ubiquitylation and deubiquitylation converts the dormant pool of inactive Met4 into an active transcription factor. Met4 together with several co-factors induces a transcriptional program to restore sulfur metabolite homeostasis and to protect from heavy metal stress. A cell cycle arrest that specifically requires Met32 is also induced to protect cellular and genomic integrity during stress. (B) The transcriptional activator Met4 is substrate and substrate adapter of the ubiquitin ligase SCFMet30-Met4 and controls transcription complex dynamics. The transactivator function of Met4 is inhibited by SCFMet30-mediated ubiquitylation. Met4 ubiquitylation does not regulate its function as substrate adapter of the ubiquitin ligase SCFMet30-Met4, which leads to ubiquitylation and degradation of the DNA-binding co-factors Met31, Met32 and Cbf1. Stress signals block SCFMet30 resulting in simultaneous activation of Met4 and stabilization of the Met4 bound co-factors to assemble the active Met4 complex. A constitutive second degradation pathway independent of SCFMet30-Met4 selectively removes unbound co-factors. Repression after disappearance of the stress signal induces ubiquitin-dependent co-factor degradation and Met4 inactivation.