Figures & data
Figure 1 Characterization of CUT and SUT non-coding transcripts from bidirectional promoters. (A) Schematic representation of a bidirectional promoter. Position of a strand specific probe to the body of the CUTs/SUTs is shown. (B–E) Northern blotting of non-coding transcripts in candidate chromatin and RNA processing mutants. CUTs and SUTs and the extended transcripts (eCUTs/eSUTs) are indicated on the left side of the blots while the positions of the 500 and 1,000 nucleotide size standards are marked on the right. The membranes were stripped and re-probed to detect the ACT1 actin mRNA loading control. Quantification of non-coding transcript species abundance relative to ACT1 is given in numbers below the images, “B“ signifies that no signal above background was observed.
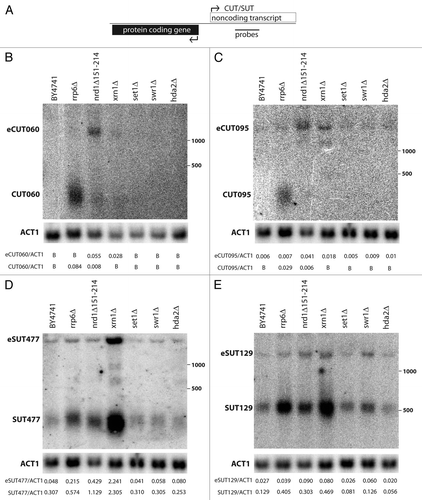
Figure 2 Stabilization of SUTs and CUTs in rrp6Δ is post-transcriptional. (A) Analysis across the bidirectional ERO1/SUT285 region. Schematic figure of the locus is at top with position of ChIP primer pairs indicated. Top graph shows ChIP signal for TBP in RRP6 (white) and rrp6Δ (gray) cells. Lower part shows ChIP for the Rpb3 subunit of RNA pol II. Data was generated by qPCR and is expressed as fold-enrichment of the indicated region relative to a non-transcribed telomeric control. Three biological and technical repeats were analyzed, error bars denote standard errors. (B) Analysis of Rpb3 across the bidirectional UTP6/CUT095 region, as in part (A).
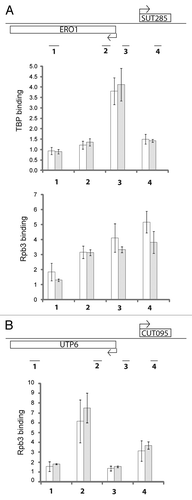
Figure 3 CUT095 and SUT129 transcript profiles in a collection of RNA decay pathway mutants. Northern blots of CUT095 (top) or SUT129 (middle) of RNA samples from the indicated RNA decay mutants are shown. The expected size transcripts and the extended eCUTs/eSUTs are indicated at left. The membranes were stripped and re-probed to detect the ACT1 (actin) mRNA as a loading control (bottom). Note that BY4741 is the isogenic “wild-type” control for strains to the left of the vertical line while W303-1B is the corresponding control for strains to the right. Quantification of non-coding transcript species abundance relative to ACT1 is given in numbers below the images, “B“ signifies that no signal above background was observed.
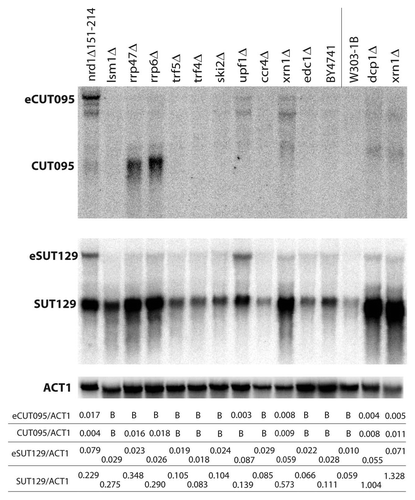
Figure 4 Characterization of 3′ extended non-coding transcripts originating from bidirectional promoters. (A) Northern blots of CUT095 were hybridized with strand-specific probes recognizing top strand transcripts as diagrammed in the schematic figure, detecting transcripts 5′ of CUT095 (P5′, left), CUT095 (P95, middle) or transcripts 3′ of CUT095 (P3′, right). Note the P95 part is the same blot that appears in . (B) Total RNA was hybridized with no oligo (-) or antisense oligos and to the regions of the SUT129 locus indicated in the diagram on top and then treated with RNAse H. The digested samples were then analyzed by northern blotting using a probe against the middle of SUT129 (gray bar). Note that the eSUT levels decrease only with the middle (M) or 3′ oligos. Quantification of the eSUT/SUT ratio of two repeats is given below, error bars denote standard errors. Note that for the M lane, the SUT signal is the combination of both smaller bands. (C) Effect of CF IA inactivation on short and extended non-coding transcripts. RNA from the indicated strains grown at permissive (25°C) or nonpermissive (37°C) temperatures were analyzed by northern blot. SUT and eSUT transcript species are indicated. The membranes were stripped and re-probed to detect the 5S rRNA (5S) as loading control (bottom). Quantification of non-coding transcript species abundance relative to 5S is given in numbers below the images, “B“ signifies that no signal above background was observed.
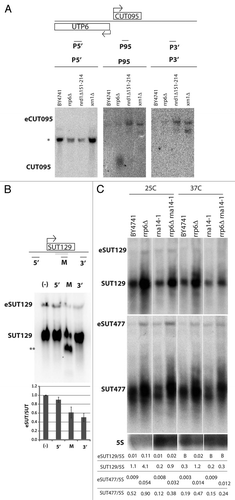
Table 1 Frequency of Nrd1 and Nab3 recognition sequences associated with CUTs and SUTs
Table 2 Comparison between estimated and expected non-coding transcript sizes