Figures & data
Figure 1. (A) In vivo chromatin organization of the HIV LTR and distribution of putative transcription factor binding sites identified using the TF consite bioinformatics tool. (B) Location of the strictly positioned HIV-1 LTR nucleosomes correlates negatively with the predicted histone binding affinity score (nucleosome score) of the DNA sequence encompassing the HIV LTR. Predicted nucleosome affinity for HIV nucleotide sequence 1–720 was determinedCitation5 using the algorithm described.Citation25 Means and standard deviations for nucleosome score at insertion sites are indicated by dashed black (mean) and gray (SD) lines and give reference to known genomic sites of HIV integration.Citation63
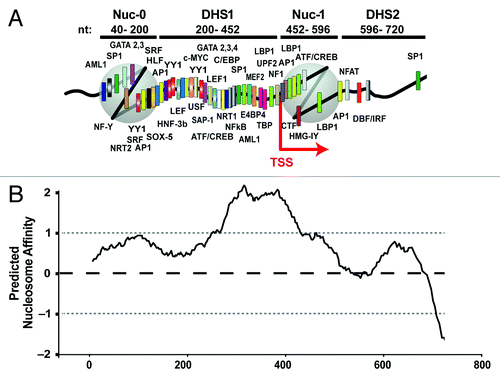
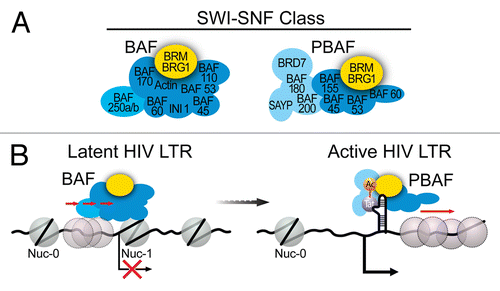
Figure 2. (A) Subunit composition of the two distinct mammalian SWI/SNF complexes, BAF and PBAF. (B) Model for SWI/SNF regulation of HIV LTR transcription. BAF actively counteracts intrinsic histone-DNA sequence preferences within HIV LTR and pulls a preferred nucleosome over DHS1 onto DNA sequences less favorable for nucleosome formation immediately downstream of the TSS, leading to positioning of nuc-1 and transcriptional repression. Upon activation, BAF dissociates from the LTR resulting in de-repression of HIV transcription and expression of Tat. Acetylated Tat selectively recruits the PBAF complex, which actively re-positions nucleosomes formed downstream of TSS enabling efficient transcription elongation.