Figures & data
Figure 1.Med23 modulates Egr1 basal transcription in vivo without changing RNAP II recruitment. (A) Time-course analysis of Egr1 expression after serum addition to serum-starved WT and KO ES cells. The expression was normalized to EF2 mRNAs, and the normalized value in serum-starved WT cells at time 0 was defined as 1. (B) Real-time PCR analysis of the Egr1 expression at steady-state and under serum starvation. The expression was normalized to EF2 mRNA expression, and the normalized value in WT cells at steady-state was defined as 1. In this and subsequent figures, results are expressed as the mean ± s.d. of n = 3 independent experiments. (C) Time-course ChIP analysis of RNAP II binding to the Egr1 promoter after serum addition. Values from real-time PCR were normalized to the percentage of input chromatin. (D) ChIP analysis of RNAP II binding to the Egr1 gene using primers for the indicated fragments. The scale is in base pairs.
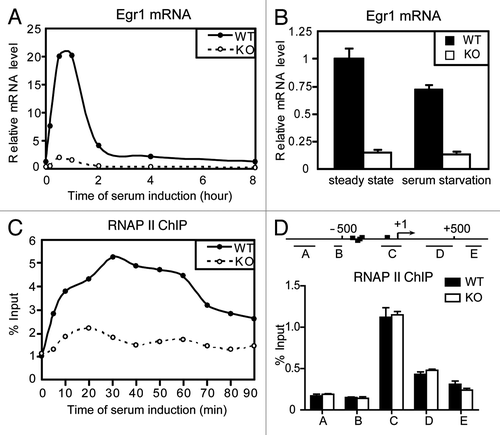
Figure 2.Med23 deficiency does not alter the occupancies of GTFs at the Egr1 promoter under an unstimulated condition. ChIP experiments were performed using antibodies against TBP (A), TFIIA (B), TFIIB (C), TFIIE (D), and TFIIH (E). The precipitated DNA was analyzed by real-time PCR with primers described in . WT+S/KO+S: 30 min after serum addition. (E) and (F), Egr1 mRNA levels in siCtrl and siTFIIEa/siCdk7 cells. The expression was normalized to EF2 mRNA expression, and the normalized value in siCtrl WT cells was defined as 1.
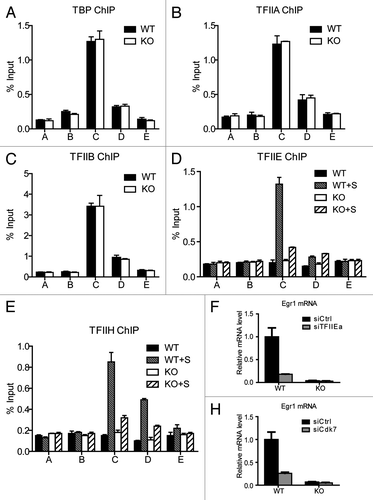
Figure 3. ELK1 and Mediator occupancies at the Egr1 promoter are not affected by the presence or absence of Med23. ChIP experiments were performed using antibodies against ELK1 (A), p-ELK1 (B), the Mediator Complex (Med1 and Med17) (C), and CDK8 (D). (E) KO MEF cells were infected with retroviruses encoding hMED23 and subsequently selected for hygromycin resistance. Western blotting was used to detect the protein levels of MED23 in WT, KO, and KO+Med23 cells. The anti-TBP blot was included as an internal control. (F) Real-time PCR analysis of the Egr1 expression in states of serum starvation and serum induction (30 min). The expression was normalized to EF2 mRNA expression, and the normalized value in serum-starved WT cells was defined as 1.
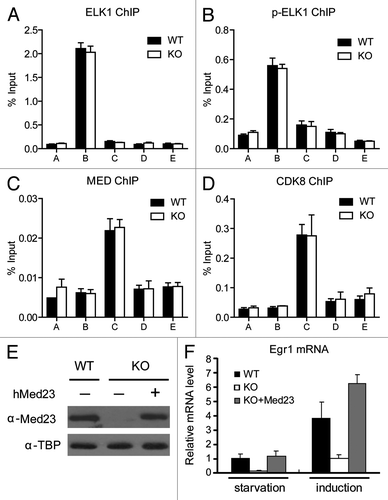
Figure 4. The interaction between ELK1 and Med23 is necessary for maintaining Egr1 basal transcription. (A) The MEK1/2 inhibitor U0126 was added to the culture medium for 30 min, and total RNA samples were harvested and analyzed by real-time PCR. (B) WT and KO ES cells were treated or not treated by U0126 for 30 min. ChIP assays were performed using RNAP II antibody. (C) The structures of Gal4-Elk1 and Gal4-AA (S383,389A) are schematically depicted. The sequences are shown below. D: D-box domain; C: C-box domain. (D) 293T cells were transfected with a 5 × Gal-E1B-TATA-luciferase reporter construct and a plasmid encoding the Gal4-Elk1 activation domain or Gal4-Elk1 mutant, with a MEKK expression plasmid. Firefly luciferase activity was normalized to Renilla luciferase activity.
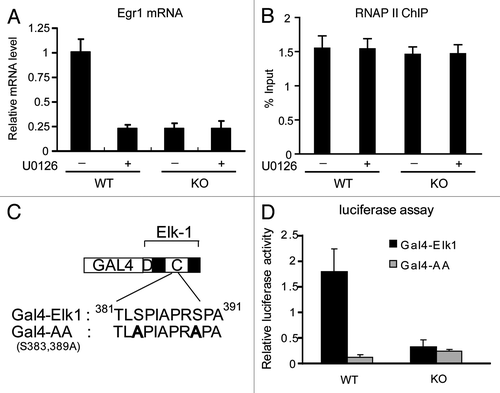
Figure 5. MED23 is required for RNAP II CTD phosphorylation and RNAP II elongation but not RNAP II recruitment. ChIP experiments were performed using antibodies against total RNAP II (A), phosphor-Ser5 CTD (Ser5P) (B), and phospho-Ser2 (Ser2P) (C) at the Egr1 locus using primers as indicated below.
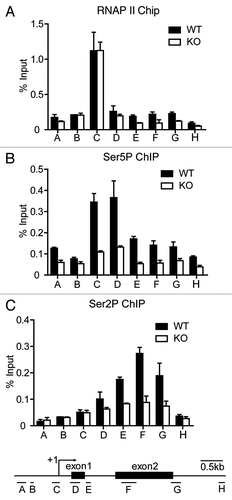
Figure 6. MED23 is partially required for the recruitment of CDK9 to the Egr1 promoter. ChIP experiments were performed using antibodies against NELF-E (A), Spt5 (B), and CDK9 (C) at the Egr1 locus, as described in . (D) Egr1 mRNA levels were measured by real-time PCR in WT and KO ES cells with or without 1 h of flavopiridol (250 nM) pre-treatment. The expression was normalized to the EF2 mRNA expression. (E) WT and KO ES cells were infected with retroviruses encoding CDK9. Stable cell lines were selected for hygromycin resistance, and the CDK9 protein level was analyzed by western blotting. β-actin was used as an internal control. (F) Egr1 mRNA levels were measured by real-time PCR in control- and CDK9-overexpressed WT and KO ES cells. (G) ChIP analysis using an antibody against CDK9 at the Egr1 locus in control- and CDK9-overexpressed WT and KO ES cells. The mean of at least three separate experiments is shown, and the standard deviation is indicated. Student’s t test, **p < 0.001.
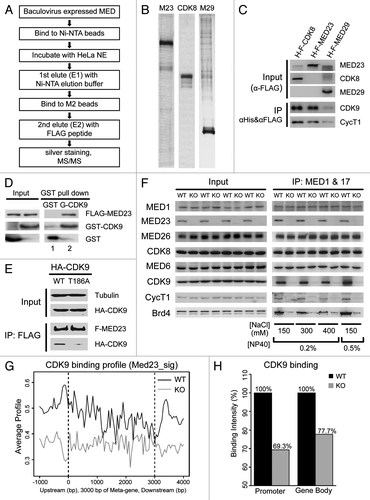
Figure 7. MED23 interacts with CDK9 in vitro and in vivo. (A) Scheme for tandem affinity purification and MS/MS to identify Mediator subunit binding proteins. (B) Baculovirus-expressed His-Flag-MED23 (M23), His-Flag-CDK8 (CDK8) and His-Flag-MED29 (M29) were purified by Ni-NTA beads and then FLAG M2 beads sequentially, as described in (A). The proteins eluted from FLAG M2 beads were analyzed by SDS- PAGE followed by silver staining. (C) The proteins eluted from FLAG M2 beads were analyzed by SDS- PAGE followed by immunoblotting with antibodies against CDK9 and Cyclin T1. (D) Soluble His-Flag-MED23 was incubated at 4 degree overnight with immobilized GST or GST-CDK9. After washing, the bound proteins were eluted by boiling and immunoblotted with the indicated antibodies. (E) Flag-Med23 plasmid was co-transfected with HA-cdk9 or its mutant HA-cdk9 (T186A) into 293T cells. Whole-cell extracts were used for immunoprecipitation with the anti-FLAG M2 beads, followed by immunoblotting using antibodies against Tubulin, FLAG or HA. (F) Nuclear extracts prepared from wild type (WT) or Med23−/− (KO) mouse embryonic stem cells were subjected to co-IP with MED1 and MED17 antibodies under various stringent wash conditions as indicated. The immunoprecipitated proteins were detected with indicated antibodies by immunoblotting. (G) ChIP-seq analysis of CDK9 enrichment on the set of genes whose expression level are regulated by MED23, in WT (black) and KO (gray) ES cells. All these genes were normalized to 3 kb for Mata-gene, with 1 kb extended upstream from TSS, and 1 kb downstream from TTS for analyzing average profile in 50 bp bins. (H) Quantitation of CDK9 binding intensity on promoter region (1 kb upstream from TSS) and gene body region (from TSS to TTS). Both are normalized to the values derived from the WT ES cells.