Figures & data
Figure 1. The effect of CAP operator position at OL on activation of PRM. (A) A CI octamer-mediated complex which bridges OR and OL forms a 2.3kb DNA loop, and allows the α subunit of an RNAP (αCTD) bound at PRM to contact the UP element at OL. (B) A structural model of an RNAP-lambda CI-OR-OL complex, assembled as described in Cui et al.,Citation16 and positioned to mimic the view in (A). The model shows that when an octamer of CI (red) connects OR and OL, it is plausible that an RNAP αCTD subunit (green) can interact with the UP element sequence (blue) at OL. The DNA connecting OR and OL is represented by a red line. (C)The α subunit of RNAP is known to interact with DNA-bound CAP.Citation30 This cartoon shows how a CAP operator, artificially replacing the UP element, may be able to recruit an RNAP α subunit to OL, and thereby activate PRM. We predicted that this contact, and hence the degree of PRM activation, would be sensitive to CAP operator sp.acing. (D) A structural model of an RNAP-lambda CI-DNA-CAP-αCTD subunit complex was constructed by replacing the UP element DNA in part (B) with the CAP-DNA-αCTD structure (taken from PDB 1LB2). The CAP-DNA-αCTD complex has been positioned to reproduce the CAP-10 construct (below), where the edge of the CAP operator is positioned 10bp from the edge of the OL3 operator. The DNA connecting OR and OL is represented by a red line. Inherent flexibility of the OL DNA, the CI octamer complex and the αCTD linker peptide (shown as a green dashed line) means that the structure shown here is one of many possible conformations of the complex. (E) The UP element at OL was replaced with a consensus operator for the CAP protein. The arrangement of the PRM lacZ reporters used to assay the effect of CAP operator position relative to OL is shown. A CAP consensus operator was placed at various positions (distances in base pairs are indicated) on the OL3 side of the distal OL site, replacing the λ UP element. OR3− and OL3− mutations were used to eliminate CI repression of PRM. (F) Effect of CAP site sp.acing on CI activation of PRM. The 3.3 WLU λCI expression construct was integrated at φ 186 attBCitation16 and CAP was expressed from the wild type chromosomal gene. CI activation is defined as the fold increase of PRM activity at 3.3 WLU CI over basal (absence of CI). Error bars represent 95% confidence intervals (n ≥ 8). The mean activity of PRM in the absence of CI was 143 ± 5 units (n = 56).
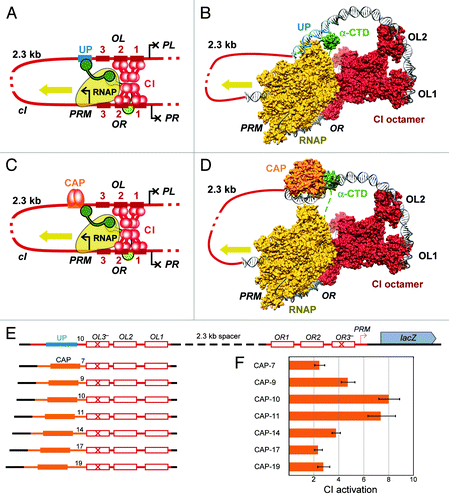
Figure 2. Lambda OL as a multi-faceted enhancer. The types of long-range protein-protein and protein-DNA interactions that characterize an RNA polymerase-enhancer complex are depicted (A), and compared with the corresponding interactions that occur during transcription from the lambda PRM promoter (B).