Figures & data
Figure 1. Analysis of ChIP-seq and ChIP-chip data reveals differences in spacing distributions of GAS-like motifs for different forms of STAT proteins. (A) IL-2 induced WT P-STAT5A favors two GAS motifs separated by 11–12 bp (mm8 data set, GSE36890), while this spacing of 11–12 bp is absent for DKI P-STAT5A that prohibits tetramer formation in mouse T cells (B). (C) Analysis of LIF-induced P-STAT3 binding in mouse ES cells shows the spacing distribution between two GAS-like motifs with significant peaks at 20 bp and 24 bp (mm8 data set, GSM288353). (D) Analysis of STAT3 binding in DU145 prostate cancer cells with low levels of P-STAT3 shows the spacing distribution peaked at about 50 bp, 70 bp, 90 bp, and 140 bp (hg18 data set, GSE25866). Genome-wide distributions of GAS binding sites bound by various STATs indicate that only 8–12% of all GAS sites are located in the promoter regions with the majority in introns and intergenic regions for both mouse genome (mm8) and human genome (hg18).
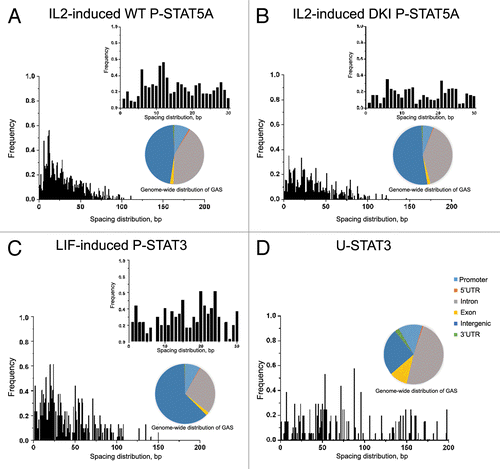
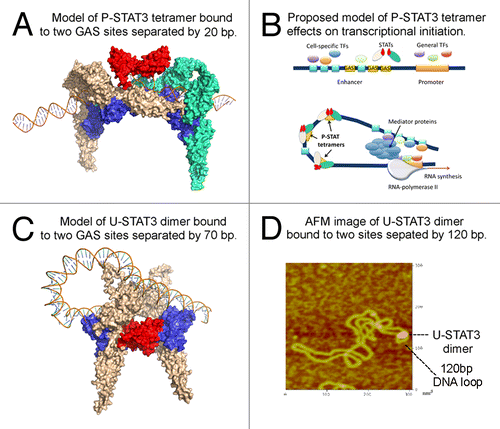
Figure 2. STAT3 may regulate chromatin topology through bending and looping of DNA. (A) Modeled structure of phosphorylated STAT3 tetramer bound to DNA. Protein structure was generated using I-TASSER. The DNA-protein complex structures are constructed using 3dRNA and 3dRPC programs designed for structural modeling and docking of nucleic acid structures. The two dimers (colored in wheat and green, respectively) are linked via ND-ND interaction (colored in red) and they bind to DNA via DBD (colored in blue). (B) Proposed mechanism by which P-STAT3 tetramer may regulate gene expression through DNA bending. General transcription factors bind to promoter region, while tissue-specific and/or inducible transcription factors bind to their cognate binding sites within an enhancer sequence. In order for enhanceosome to be assembled, these transcriptional factors need to be positioned in the optimal way. This may be achieved through DNA bending caused by DNA-bending proteins, in particular P-STAT3 tetramer (or a series of P-STAT3 tetramers), to promote cooperation between various proteins bound to distant DNA regions. Once all players are interacting with each other, mediator complex, and RNA-polymerase II, the transcription is initiated. (C) Modeled structure of U-STAT3 dimer bound to two DNA sites separated by 70 bp suggests that U-STAT3 dimer may regulate gene expression through DNA looping. The STAT3 ND is colored in red and the DBD is colored in blue. (D) Atomic force microscopy image of U-STAT3 dimer bound to DNA forming a loop of about 120 bp. The data was generated using characterized supercoiled pUC8F14C plasmid and purified full-length recombinant unphosphorylated STAT3.