Figures & data
Figure 1. A model of sequence-specific pausing. (A) Pause-free elongation. RNA (orange), template DNA strand (gray), catalytic Mg2+ (magenta circle), and two RNAP domains (blue) involving 5′ RNA separation from the hybrid, i.e., Switch 3 (arrow head) and lid (triangle) domains are shown. The 3′ RNA-binding site (i) and the NTP binding site (i+1) are also indicated. (B) Elongation at the pausing site. The two sequence elements involved in transcription pausing are shown: (1) 3′ ACGC 5′ sequence in the transcribed DNA strand (grey) corresponding to the junction between the RNA-DNA hybrid and the downstream dsDNA in the elongation complex (indicated by shaded box); this sequence increases mobility/flexibility of the RNA/DNA backbones, which promotes fraying of the 3’ RNA end. (2) G residue in the RNA at the upstream end of the hybrid contributes to immobilization of the hybrid in the catalytic cleft of RNAP by interacting with the Switch 3 domain in the post-translocated state, or by interacting with the lid domain in the pre-translocated state.
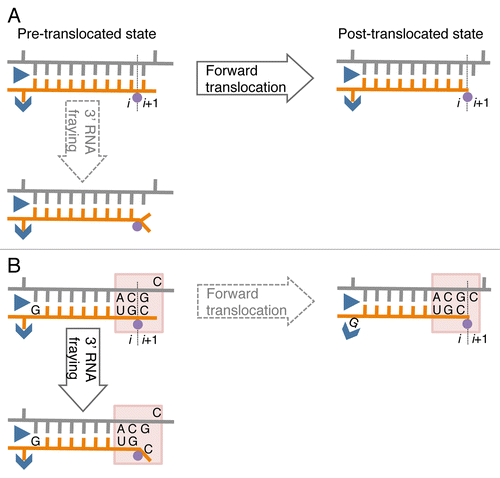
Figure 2.Cis- and trans-acting factors affecting translocation. (A) Structure of RNA-DNA hybrid and dsDNA in TEC by T. thermophilus (Tth) RNAP. The structural targets for translocation regulators are indicated by arrows. (B) A schematic structure of TEC: RNAP (blue oval), upstream and downstream dsDNA (gray cylinders), RNA-DNA hybrid (brown cylinder), transcription bubble (black line) and the bridge helix (green) are shown; the active center in RNAP is represented by a circle with i and i+1 subsites. The inset displays the pre-translocated configuration of the active center with DNA lesion in i site (yellow triangle) and the 3′ RNA residue in a frayed configuration in i+1 site. The left side shows cis-acting translocation inhibitors: bending, or other structural alteration of the hybrid, the front-end DNA duplex and hairpin in the nascent RNA interacting with RNAP (shown by curved arrow). The right side displays the trans-acting inhibitors: a drug molecule bound to bridge helix reducing its mobility/bending (red dot), protein factors bound to dsDNA, nascent RNA or RNAP, and the second RNAP molecule in a head-to-tail configuration (all in magenta).
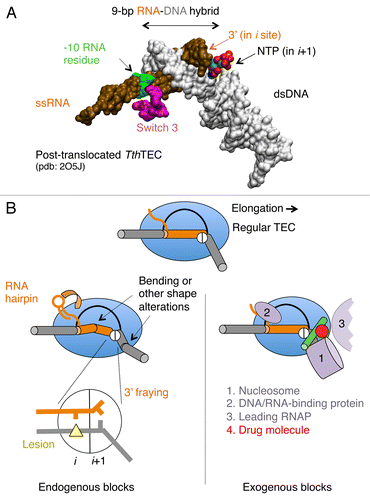
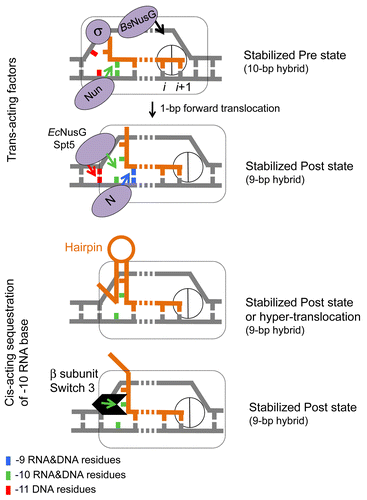
Figure 3. Translocation modulators target RNA-DNA hybrid and transcription bubble in bacterial TEC. Top panel: Nun protein of bacteriophage H022 interferes with translocationCitation68 by stabilizing the -10 base pair of the hybrid and tethering RNAP to the hybrid. We proposed that the homologous N protein of bacteriophage λ stabilizes the -9 base pair of the hybrid and prevents the -10 base pair to favor translocation. E. coli NusG and its eukaryotic homolog Spt5 promote translocation by facilitating re-annealing of DNA immediately upstream from the 9-bp hybrid. B. subtilis NusG tethers RNAP to pre-translocated register by binding to the middle part of transcription bubble.Citation70 σ70 subunit interferes with translocation by binding to the “-10-like” sequence at the upstream end of transcription bubble.Citation96 Bottom panel: cis-acting RNA hairpin and Switch 3 domain in β subunit promote translocation by preventing expansion of the hybrid to 10-bp length.Citation79