Figures & data
Figure 1. mRNP formation and export to the cytoplasm. Several mRNA binding proteins decorate the nascent mRNA forming an mRNP. One of the protein complexes coupling transcription to export of the mRNP is the TREX complex. The Prp19 complex (Prp19C) is essential for the occupancy of TREX at transcribed genes. The C-terminus of the Prp19C subunit Syf1 is necessary for efficient recruitment of Prp19C and TREX. After cleavage and polyadenylation the mRNP is released from the site of transcription and remodeled to an export competent mRNP. The export receptor Mex67-Mtr2 is recruited to the mRNP via direct interaction between the TREX component Yra1 and the mRNA export receptor. In addition, Hpr1, Nab2, Npl3 and the THSC-complex are implicated in Mex67-Mtr2 recruitment to the mRNP. Mex67-Mtr2 then exports the mRNP through the nuclear pore complex. Most S. cerevisiae proteins depicted here are conserved in higher eukaryotes.
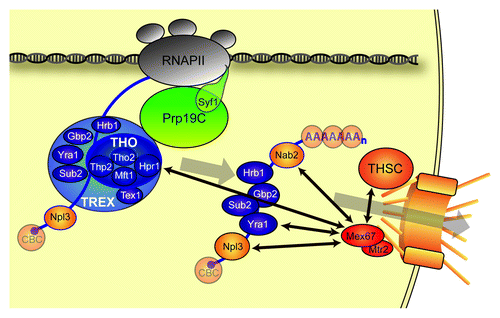
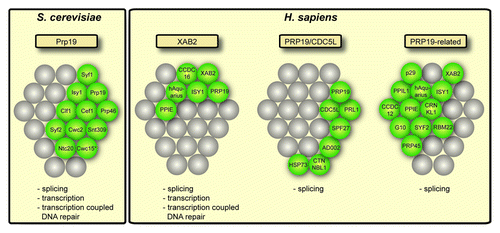
Figure 2. Conservation of the Prp19 complex (Prp19C). In S. cerevisiae, one Prp19C has been identified that consists of ten subunits. In higher eukaryotes, e.g. H. sapiens, three different Prp19 complexes have been identified: The XAB2 complex, the PRP19/CDC5L complex and the PRP19-related complex. The proteins present in each complex are depicted in green. The position in the grid indicates the homology between S. cerevisiae and H. sapiens proteins. Cwc15 in S. cerevisiae (indicated by a star) has not been found in yeast Prp19C so far, but is homologous to human AD002 and was found to physically interact with yeast Clf1, a subunit of yeast Prp19C. Below each complex the known functions of each complex are given.