Figures & data
Table 1. BET family proteins
Figure 1. Phenotypes of bet-1 mutants in C. elegans. (A) The T-cell lineage in wild-type (WT) and bet-1 mutants. The posterior granddaughter cells show abnormal transformation of cell fate from neuroblasts to hypodermal cells. (B) Schematic drawing of bet-1 phenotype in the posterior lateral ganglia. In the wild-type V5.pa lineage, the PDE and PVD neurons express osm-6::gfp and dop-3::rfp, respectively. In bet-1 mutants, abnormal transformation from the PDE fate to the PVD fate was observed. (C) In the Z1/Z4 lineage, which produces the somatic gonad, wild-type animals produce two distal tip cells (DTCs) at the late L1 stage. In bet-1 mutants, abnormal transformation to the DTC fate occurs in the late larval stages, leading to the production of extra DTCs.
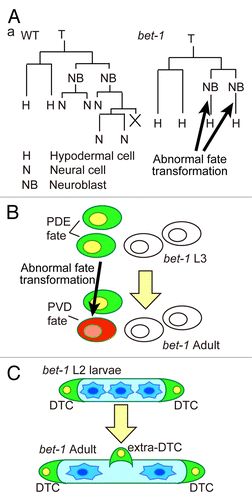
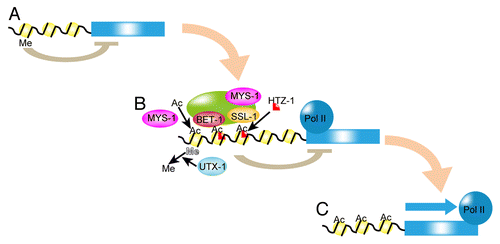
Figure 2. Working model for HTZ-1-dependent cell fate maintenance. (A) H3K27me silences the selector genes (blue box). Me, black line and yellow circle are methylation on H3K27, DNA, and histone, respectively. (B) UTX-1 demethylates H3K27me on selector gene loci. MYST histone acetyltransferases, including MYS-1 and MYS-2, acetylate the selector gene loci. Then, SSL-1 in the BET-1-containing complex deposits HTZ-1, which represses the transcription of selector genes. HTZ-1 may maintain the poised state of RNA polymerase II (Pol II). The transcription of selector gene(s) is ready to be activated. (C) When a cell receives a differentiation signal, repression by HTZ-1 is released and the gene is transcribed.