Abstract
Background
Nucleophosmin/B23 (NPM) is a 38 kDa molecular phosphoprotein involved in ribosome assembly and transport. Findings have revealed a complex scenario of NPM functions and interactions, pointing to proliferative and growth-suppressive roles. NPM appears to be more abundant in tumour cells than in normal cells.
Objective
The aim of the work was to identify cytoplasmic localization of NPM using bone marrow clot biopsy and correlate it with disease and patient characteristics and the known prognostic factors, induction chemotherapy response and survival after 12 months of follow up.
Methods
The present work was undertaken on 50 cases of normal karyotype acute myeloid leukaemia classified according to modified FAB system. Bone marrow aspirates were obtained, clotted and a formalin-fixed, paraffin embedded cell block was prepared. Sections were immunostained for NPM.
Results
Twenty-six cases (52%) had cytoplasmic positivity (cNPM+) while the remaining 24 cases (48%) had nuclear restricted NPM (cNPM−). cNPM positivity was significantly variable among the different FAB subtypes being the most prevalent among M5 and M2 cases, was associated with high blast and white blood cell count at diagnosis. It was significantly correlated with CD34 negativity, CD14 positivity but not with FLT3 mutations. Clinically no relation between cNPM positivity and age, sex nor extramedullary involvement of the studied patients was proven. The cytoplasmic positivity for NPM was significantly correlated with increased survival and better outcome after cycles of chemotherapy. So it can be regarded as a good prognostic marker. FLT3 positivity affected the survival of the cNPM negative group of patients remarkably, denoting the importance of combining both their statuses to predict outcome of therapy and survival.
Conclusion
Our data confirm that cytoplasmic NPM1 immunoreactivity is predictive of NPM1 mutations and can be included in the routine diagnostic and prognostic workup of AML.
1 Introduction
Acute myeloid leukemia (AML) is clinically, cytogenetically, and molecularly heterogeneous.Citation1 About 30% of cases carry recurrent chromosomal abnormalities that identify leukemia entities with distinct clinical and prognostic features,Citation2 and 10–15% of AMLs have nonrandom chromosomal abnormalities. In cytogenetic analysis, 40–50% of AMLs show normal karyotype (AML-NK) and, biologically and clinically, are the most poorly understood group.Citation1,Citation3,Citation4 Attempts to stratify AML-NK using gene microarrays succeeded in associating gene-expression patterns with differences in response to treatment, but no specific genetic subgroups emerged.Citation5,Citation6
Table 1 The correlation between the AML subtype and cytoplasmic expression of NPM (cNPM).
Molecular analyses of AML-NK identified several mutations that target genes encoding for transcription factors (AML1 and CEBPA: 2–3% and 15–20% of cases, respectively),Citation7–Citation10 receptor tyrosine kinases (FLT3 and KIT: 25–30% and 1%, respectively),Citation11–Citation13 and the RAS genes (10%),Citation14 as well as a partial tandem duplication of MLL gene (MLL-PTD: 5–10%).Citation15–Citation17
Our understanding of acute myeloid leukemia (AML) has improved dramatically in the past years with the discovery by Falini et al.Citation18 that Nucleophosphamin 1 (NPM1) is mutated in around half of cases with a normal karyotype, making it the most common molecular lesion identified in this disease to date.
Nucleophosmin is a highly conserved phosphoprotein that is ubiquitously expressed in tissues.Citation19 It is one of the most abundant of the approximately 700 proteins identified to date in the nucleolus by proteomics.Citation20,Citation21 Although the bulk of NPM resides in the granular region of the nucleolus,Citation22,Citation23 it shuttles continuously between nucleus and cytoplasm,Citation23,Citation24 as proven by interspecies heterokaryon assays. NPM nucleocytoplasmic traffic is strictly regulated, since important NPM functions, including transport of ribosome components to the cytoplasm and control of centrosome duplication, are closely related to its ability to actively mobilize into distinct subcellular compartments.Citation25
The NPM1 gene, mapping to chromosome 5q35 in humans, contains 12 exons.Citation26 It encodes for 3 alternatively spliced nucleophosmin isoforms: B23.1, B23.2, and B23.3.Citation27,Citation28
Functions of NPM1 are not completely understood, it is thought to play an important role in centrosome assembly, and has RNA binding and chaperone activity. It regulates the Arf-p53 pathway, suggesting that NPM1 has tumor-suppressor activity, a hypothesis supported by murine models of NPM1 loss of function.Citation29
NPM1 mutations occur in 50–60% of adult acute myeloid leukemia with normal karyotype (AML-NK) and generate NPM mutants that localize aberrantly in the leukemic-cell cytoplasm, hence the term NPM-cytoplasmic positive (NPMc AML).Citation30
The various NPM1 mutations identified in AML are heterozygous and involve the C-terminal region encoded by exon 12. These not only disrupt key tryptophan residues that are required for localization to the nucleolus, but also generate a nuclear export signal leading to delocalization of mutant NPM1 to the cytoplasm where it sequesters residual wild-type protein from the nucleus. As an alternative to sequence analysis, the presence of an underlying NPM1 mutation can be inferred by immune-histochemistry, which shows an abnormal cytoplasmic localization of the protein in leukemic blasts.Citation31
NPM1 mutations are reliably identified by various molecular biology techniques,Citation32,Citation33 which also allow simultaneous detection of NPM1 and FLT3-ITD or CEBPA mutationsCitation34,Citation35 (all prognostically relevant in AML-NK).Citation10
Immunohistochemical detection of cytoplasmic NPM in AML is predictive of NPM1 mutations.Citation36 The main advantages of immunohistochemistry are simplicity, low cost, and ability to correlate cytoplasmic NPM with morphology and topographic distribution of leukemic cells. It is the first-choice technique for detecting NPM1 mutations in cases of dry-tap or small biopsies (e.g., skin involvement in myeloid sarcoma),Citation37 and also helps rationalize cytogenetic/molecular studies in AML.Citation36
2 Methods
The present study was conducted on 50 newly diagnosed (de novo) patients with Acute Myeloid Leukemia with normal karyotype of all FAB subtypes except M3. Subjects were taken from the Medical Research Institute hospital in Alexandria. Consent of the patient was required for participation in the study.
Cases of AML were taken consecutively, only adult cases were included, i.e., 18 years and above, both sexes were included in the study. AML secondary to MDS, AML therapy related, and cases that have received any type of chemotherapy were excluded.
The treatment schedule for newly diagnosed patients consisted of induction cycle(s) using the (3 + 7 protocol) consisting of a combination of Cytosine arabinoside (Ara C), and an anthracyclin. Patients less than 50 years of age received late consolidation cycles with a high dose Ara-C regimen with or without novantrone. Patients more than 50 years received either low dose or high dose regimens for consolidation according to their general conditions. Some patients above 60 years of age did not take any chemotherapy along the course of the disease; only supportive measures were administered to them. Patients not responding to 2 cycles of the conventional (3 + 7) protocol were treated with high dose Ara-C and mitoxantrone (novantrone) (HAM regimen) and were then filed for bone marrow transplant if possible.
2.1 Follow up of patients
For all patients in the study, a follow up of 12 months was included to detect the effect of cNPM on response to therapy. Cases that died during induction therapy, or cases who did not receive chemotherapy were non-evaluable cases and were excluded from the follow up study.
2.2 Methods
All newly diagnosed AML cases in the study were subjected to:
Detailed history taking and complete clinical examination.
Laboratory investigations including:
| (a) | Diagnostic investigations:
| ||||||||||||||||
| (b) | Routine investigations: For all patients the following was done at diagnosis: Blood chemistry including liver function test (AST, ALT),Citation42 serum uric acid, kidney function tests (urea and creatinine),Citation43 serum albumin and LDH.Citation43 ESR and C-reactive protein. CSF examination for blast cells and culture, ultrasonographic examination of the abdomen and chest X-ray were done to all patients. | ||||||||||||||||
| (c) | Conventional cytogenetics: Cytogenetic study was done; the karyotype of the blast cells was determined by banding techniques to detect visually accessible aberrations. G banding either direct or after 24 h culture in RPMI without stimulation with methylacetic acid fixation was done to cases. Cases with any visible abnormality in karyotype were excluded from the study.Citation44 | ||||||||||||||||
| (d) | FLT3 mutational status was retrieved from patients’ records. | ||||||||||||||||
2.3 Morphologic examination and NPM immunostaining of bone marrow clot biopsyCitation18,Citation45
Cell blocks of bone marrow clot biopsy obtained from each patient were processed. They were routinely fixed in 10% neutral buffered formalin and embedded in paraffin. Tissue sections (4 μm) were routinely stained with Haematoxylin and Eosin (H&E) stain to confirm the diagnosis.
Similar sections were deposited on SuperFrost Plus Slides (Menzel-Gläser, Braunschweing, Germany). Immunostaining was performed using a standard avidin-biotin-peroxidase complex method. Antigen retrieval was accomplished by boiling in 10 mM citrate buffer, pH 6 for 10–20 min. The slides were incubated with anti-NPM antibodies (mouse monoclonal immunoglobulin (Ig)G/κ; NA24; ready to use; Lb Vision Corporation, USA). Negative controls included omitting the primary antibody and substituting it with a normal mouse IgG. A section of NPM positive chronic tonsillitis was used as a positive control.
Interpretations were performed blind to patient background information. The sub cellular distribution of NPM (i.e., restriction to the nucleus or presence in the cytoplasm) was assessed without knowledge of the FAB subtype, or molecular and immunophenotyping characteristics. Cases were classified as either NPMc+ (positive for cytoplasmic NPM) or NPMc− (negative for cytoplasmic NPM). No grading system for the intensity of immunostaining was adopted.
The aim of the present work is to identify the presence of cNPM in 50 primary diagnosed AML cases with normal karyotype. The NPM cytoplasmic localization is correlated with: the FAB typing, the known prognostic factors, induction chemotherapy response and survival after a follow up period of 12 months.
2.4 Methods of statistical analysis
After data entry into a specially designed sheet using Microsoft Excel, a print out of the data was thoroughly revised and data entry mistakes were corrected. Then the file was transferred into Statistical Package for Social Science (SPSS) version 17 format and data explore was carried out. Descriptive statistics were carried out for scale variables and they were compared using independent Student's t-test, while categorical variables were tested for association using cross-tabulation with χ2 testing. Survival analysis using Kaplan–Meier statistics and comparison of factors was carried out by Log Rank (Mantel–Cox). Survival and hazard functions were also plotted with cytoplasmic NPM as a comparison factor. The study adopted a 0.05 level of significance (alpha error) and beta error was set to be 20%.
3 Results
3.1 Characteristics of the total cohort of AML patients
The present study included 50 newly diagnosed adult patients with de novo AML. The ages of the patients ranged from 19 to 85 years with a mean of 52.44 ± 17.96 years. Of all AML patients in the study, 31 (62%) were females and 19 (38%) were males. Sixteen cases (32%) cases show extra-medullary infiltrations.
3.2 FAB classification
3.2.1 Morphologic examination
Morphologic features of the neoplastic population in bone marrow aspiration cell blocks helped by the peripheral blood film, flow cytometry and genetics results classified the studied cases accurately into the following subtypes:
3.2.1.1 Undifferentiated M0, 6 cases (12%)
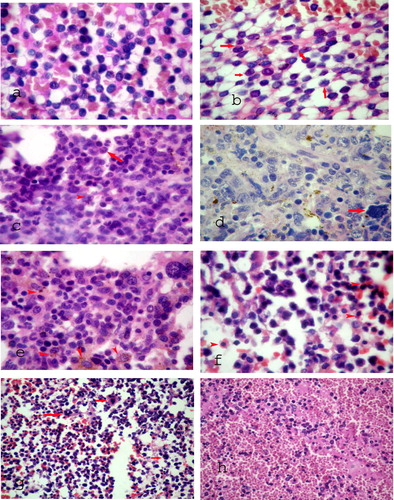
These were cases showing no signs of maturation (a).
3.2.1.2 M1 AML with minimal maturation, 9 cases (18%)
“Thumbprinting” – an indentation of the nuclear contour is noted (b).
3.2.1.3 M2 AML with maturation, 6 cases (12%)
Monocytes comprise <20% of bone marrow cells (c).
3.2.1.4 M4 acute myelomonocytic leukaemia, 5 cases (10%)
Acute leukemia with differentiation along both myeloid and monocytic lines (d).
3.2.1.5 M5 AML (monocytic), 20 cases (40%)
Large monoblasts and promonocytes with abundant cytoplasm. Granulocytes <20% of cells (e).
3.2.1.6 M6 acute erythroid leukaemia, 1 case (2%)
In acute erythroid leukemia, erythroid elements make up more than 50% of the cells in the bone marrow (f).
3.2.1.7 M7 acute megakaryoblastic leukaemia, 2 cases (4%)
AML with >30% megakaryoblasts (g).
3.2.1.8 Biphenotypic acute leukaemia (BAL) 1 case (2%)
Acute leukemias having features characteristic of both the myeloid and lymphoid lineages and for this reason are designated mixed-lineage, hybrid or biphenotypic acute leukemias (BAL) (h).
3.2.2 Haematologic profile
For the total cohort of AML patients, the hemoglobin concentration ranged from 3 to 10 g/dl with a mean of 6.44 ± 1.50 g/dl. The platelet count ranged from 2 × 109 l−1 to 91 × 109 l−1 with a mean of 20.46 ± 23.458 × 109 l−1, while the total leukocytic count ranged from 1.2 × 109 l−1 to 222 × 109 l−1 with a mean of 71.116 ± 51.895 × 109 l−1. The percentage of the bone marrow blasts ranged from 22% to 98% with a mean of 59.16 ± 24.73%.
3.2.3 Analysis of NPM mutation
3.2.3.1 Immunostaining of NPM
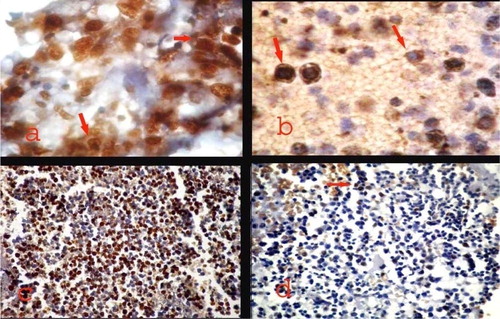
A variable pattern and subcellular localization of NPM immunostaining was noted among the 50 newly diagnosed AML patients with normal karyotype included in the study. A cytoplasmic expression (NPMc+) with or without nuclear labeling was identified in 26 cases (52%) (a). The NPMc+ cases were easily recognized since they exhibited strong cytoplasmic expression of NPM in most leukemic cells (b). Nucleus-restricted NPM expression (NPMc−) was detected in 24 AML cases (48%). These NPMc− AML specimens exhibited either heterogeneous or homogenous diffuse strong nuclear expression although in some cases nucleolar labeling became evident (c and d).
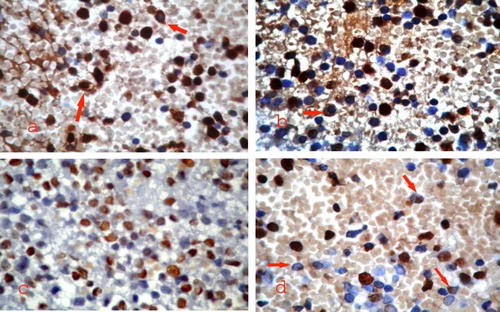
3.2.3.2 Correlation between subcellular localization of NPM immunostaining and acute myeloid leukaemia subtype ( and )
Cytoplasmic NPM (NPMc+) was predominantly observed in M2, M5a and M5b categories (15,4%, 19.2% and 46.2%) with either cytoplasmic alone or cytoplasmic in addition to the nucleus (a). M0, M1 and M4 categories showed predominant normal nucleus restricted pattern of immunostaining, and few cases showed cNPM (b). BAL and M6 cases had only an nNPM pattern (c and d).
Thus though most FAB subtypes expressed cytoplasmic NPM in variable ratios, yet statistically, a significant difference between the different FAB subtypes and cytoplasmic NPM expression was noted (χ2 = 21.125, with P value of 0.007).
White blood cell count in the NPMc negative group ranged from 1.2 × 109 l−1 to 120 × 109 l−1 (mean = 55.08, standard deviation = 394.50) while in the NPMc positive group ranged from 1.8 × 109 l−1 to 222 × 109 l−1 (mean = 88.48, standard deviation = 58.65), this was a significantly positive relation associating higher WBCs to cNPM negativity (t = 2.379, P = 0.021), and blast percentage in the NPMc negative group ranged from 22% to 90% (mean = 67.58, standard deviation = 24.99) while in the NPMc positive group ranged from 25% to 98% (mean = 51.38, standard deviation = 22.20), this was a significantly positive relation associating higher blast percent to cNPM positivity (t = 2.426, P = 0.019) were significantly higher within the cNPM negative group than the cNPM positive group, whereas no significant difference was noted as regards sex, age, platelet count, nor hemoglobin level between the studied groups. CD34 negativity was significantly highly associated with cNPM expression (χ2 = 15.751, P = 0.000), CD14 positivity was significantly associated with cNPM expression (χ2 = 9.782, P = 0.002), whereas no significant relation was detected between either FLT3 mutations (χ2 = 2.891, P = 0.84) () or extramedullary manifestations with cNPM positivity in the AML studied cases.
Table 2 Relation between FLT3 positivity and cytoplasmic NPM expression.
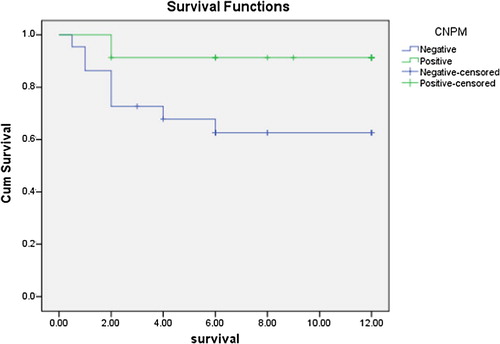
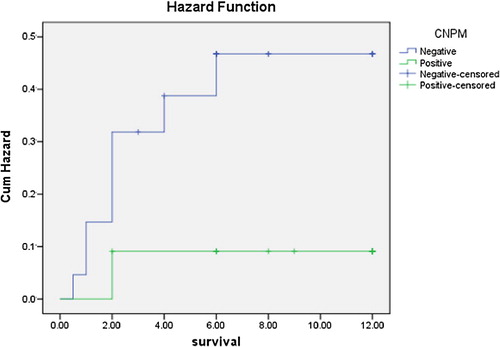
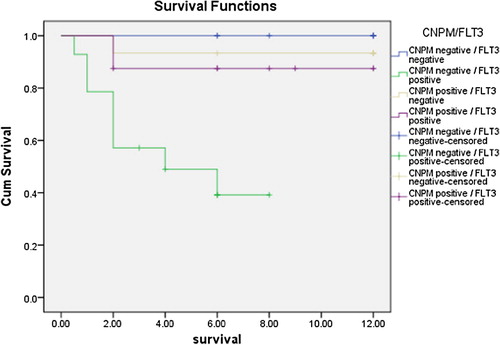
As regards outcome to therapy and survival studies, the NPMc positive group showed statistically significant higher overall survival rates in comparison to the NPMc negative group (χ2 = 5.155, P = 0.023) (), and the cumulative hazard was significantly higher in the cNPM negative group than the cNPM positive one (). The outcome of the first cycle was better for the cNPM positive group than the cNPM negative group (χ2 = 11.917, P = 0.008) (). Continuously, the outcome of the second cycle showed significantly better outcomes in the cNPM positive group than the cNPM negative one (χ2 = 17.157. P = 0.002) (). As regards FLT3 negativity was associated with statistically significant better survival after first cycle of chemotherapy (χ2 = 5.805, P = 0.018), and also the second cycle (χ2 = 8.696, P = 0.003). overall survival for the study group as regard both cytoplasmic NPM positivity and presence or absence of FLT3 mutations showed that the worst survival lied within the NPMc−/FLT3+ group and there was a statistically significant difference between this group and the NPMc+ groups regardless of their mutational status (t = 11.725, P = 0.003) ( and ).
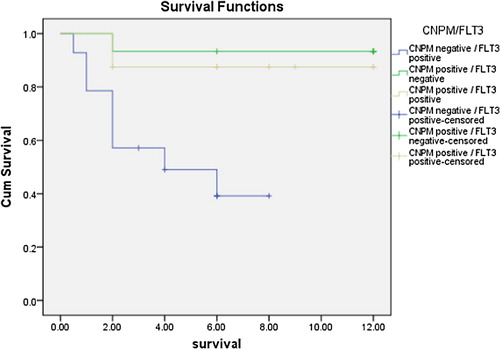
Table 3 Outcome of the first induction cycle.
Table 4 Outcome of the second induction cycle.
4 Discussion
Nucleophosmin (NPM) is a ubiquitously expressed nucleolar phosphoprotein which shuttles continuously between the nucleus and cytoplasm. Many findings have revealed a complex scenario of NPM functions and interactions, pointing to proliferative and growth-suppressive roles of this molecule.Citation46
Remarkably, heterozygous mutations of NPM are found in ∼35% of acute myeloid leukemias (AMLs), which make NPM the most frequently mutated gene in AML. Previous reports concluded that immunostaining serves as a highly specific predictor of all exon-12 NPM mutations.Citation36
The choice in the present study to detect nucleophosmin aberrant cytoplasmic expression on bone marrow clot biopsy was meant to overcome technical difficulties that appeared in previous studies, questions arose about which samples and techniques should be used. Falini et al. and Luo et al. proved that aberrant cytoplasmic expression of nucleophosmin is optimally detected in paraffin sections from B5-fixed/ethylenediaminetetraacetic acid-decalcified bone marrow trephines.Citation47,Citation48
Konoplev et al. reported less reliable results in bone marrow biopsies fixed in formalin and decalcified in formic acid.Citation49 Falini et al. preliminarily suggested that discrepancies may result from the decalcifying agent (formic acid) rather than to formalin fixation. In the methodology used in our study, clot biopsy choice was to overcome these difficulties of choosing the right decalcifying agent, and skipping the step of decalcification which leads to denaturation or change of immunogenicity of some of the proteins which leads to false results.
Consequently, use of immune staining of bone marrow clot biopsy as surrogate for molecular analysis can serve as first-line screening in AML and should facilitate implementation of the 2008 World Health Organization classification of myeloid neoplasms that now incorporates AML with mutated NPM1 (synonym: NPMc+ AML) as a new provisional entity.
In the present study cytoplasmic localization of NPM (NPMc+) was identified in 26 out of the 50 (52%) of primary AML cases with normal karyotype studied by immunostaining in bone marrow clot biopsy cell blocks. Similarly Falini et al. found NPMc+ in one third of the primary AML cases globally. In accord with our findings they had all a normal karyotype.Citation18 In another study, the same author working on AML-NK patients reported an incidence of NPM cytoplasmic positivity in 50% of these cases, approaching our results.Citation36 Moreover, Dohner et al.Citation50 and Schnittger et al.Citation51 reported an incidence of 48% and 52.9% among the normal karyotype AML cases in their study groups, respectively, which coincides with our data.
Although in the current study the presence of cytoplasmic NPM was not confined to a specific morphologic FAB subtype, interestingly, the highest frequency of NPMc+ was found in AML-M5 (17/21, 80.9%) and the lowest cases were in M1 (1/9, 11.1%) in accord with Falini et al.,Citation30 Dohner et al.Citation50 and Schnittger et al.Citation51 Falini et al.Citation30 reported that the NPMc+ pattern can be seen in AML specimens of all FAB subtypes except M4eo, and M7. However, we found the NPMc+ pattern in AML specimens of all FAB subtypes except biphenotypic leukemia and M6. Though Falini et al.Citation30 did not record the observation that cytoplasmic NPM1 is predominant in M2 specimens, we observed cytoplasmic NPM1 in high percentage in M2 (4/6 cases, 66%).
The absence of NPMc+ in M6 category found in our study goes with accordance with Zini and d’OnofrioCitation52 and it was attributed in their series to the common presence of complex karyotypes and multiple structural abnormalities in these cases reflecting their poor prognosis.Citation52 However, in all our cases normal karyotype was proven before the inclusion in the study, so the explanation of the negativity to cytoplasmic NPM still needs to be elucidated. Moreover, the relation of M6 to cytoplasmic NPM expression cannot be evaluated in our study due to the small sample study (one case) and its association with normal karyotype, thus this entity should be examined in a more detailed study including both those with normal karyotype and others with cytogenetic abnormalities whether complex or straight forward.
Dohner et al.Citation50 and Verhaak et al.Citation53 in their studies correlated a significant association between mutated NPM1 and higher WBC count, blast percentage and platelet counts. In our series, the WBC count and blast percentage were significantly higher in NPMc+ group, however, the relation with a higher platelet count was not proven.
We also confirmed that in AML-NK, the majority of NPMc+ cases were associated with the absence of CD34 expression (22/26, 84.6%), this is in accordance with Falini et al.Citation18 who found that more than 95% of NPMc+ AMLs are CD34 negative.
Consistent CD34 negativity in the great majority of studies raises the question of whether a minimal pool of CD34+/CD38− NPM1-mutated progenitors exists. In NPM1-mutated AML, investigatorsCitation54,Citation55 found that the small fraction of CD34+ hemopoietic progenitors, including CD34+/CD38− cells, carried the NPM1 mutation. When transplanted into immunocompromised mice, CD34+ cells generated a leukemia that recapitulated the patient's original disease, morphologically and immunohistochemically (aberrant cytoplasmic NPM1 and CD34 negativity).
In our series CD14 positivity was significantly associated with cNPM positivity, with the simple explanation that this marker denotes monocytic differentiation, which coincides with the high percentage of cNPM positivity with cases with monocytic leukemia.
Understanding the mechanisms leading to leukemogenesis in NPMc+ AML may lead to more specific antileukemia therapies.Citation56 NPM1 belongs to a new category of genes that function both as oncogenes and tumor-suppressor genes,Citation57 depending on gene dosage, expression levels, interacting partners, and compartmentalization. Of the most extensively studied partners of NPM1 is the FLT3-ITD. In the present studied cases, no statistically significant positive relation could be detected between FLT3 ITD and cytoplasmic NPM positivity; this can be attributed to our smaller sample size and choice of the study group to normal karyotype AML only which might led to the bias of the statistical interpretation.
The cases showing NPMc+ in the current study had a better response to induction chemotherapy, an observation that was reported by other researchers.Citation18 Recently, Konoplev et al.Citation49 showed a lack of correlation between cytoplasmic NPM1 expression and the likelihood of complete remission and prolonged survival, suggesting that NPM1 mutations, at least as detected using immunohistochemical analysis, may not be a useful prognostic indicator, though in our series the immunostaining was also the methodology used, yet overall survival was statistically significantly affected by NPMc positivity.
Furthermore, analyzing our data showed that the presence of FLT3 mutation as a single factor affected survival both after the first and second cycle of chemotherapy, and also affected the overall survival, thus prognostically the presence of a FLT3 mutation lead to shortening of the overall survival in the cohort of patients. Analyzing the combined effect of cytoplasmic NPM expression with FLT3 mutational status showed that the worst prognosis was within the NPMc−/FLT3+ group, no death occurred within the NPMc−/FLT3− group throughout the time of the study, while the NPMc+/FLT3+ and NPMc+/FLT3− groups had an intermediate survival between these 2 groups. Thus, it is apparent that the FLT3 mutational status affects the survival within the NPMc− groups more than its effect on the NPMc+ groups, its absence confers a more pronounced better prognosis within this group, than in the NPMc+ group. In accordance with our results, Hollink et al.Citation58 found similar results in their study of 297 pediatric patients with AML patients with NPM1 mutations doing significantly better than those with wild type (wt) NPM in terms of 5 year OS and EFS. However, they found that FLT3/ITD status had no significant impact on survival in patients with mutations and was only prognostically significant in patients with wt NPM1.
Controversly, Boonthimat et al.Citation59 could not observe a major difference in the overall survival (OS) in the Thai patients with and without NPM1 mutation (P = 0.376). Interestingly, statistical analyses according to the combined NPM1/FLT3 mutational status revealed a better outcome (P = 0.036) for the NPM1-mutated/FLT3 ITD-negative than NPM1+/FLT3-ITD+ and NPM1−/FLT3-ITD+ subgroup, implicating the negative impact of FLT3 mutations regardless of the NPM1 mutational status.
Thus, the detection of cytoplasmic NPM in the present work further supports the routine use of immunostaining on bone marrow aspirate cell blocks as a simple, inexpensive, and rapid method capable of first-line screening AML cases for NPM gene mutations and to rule out recurrent chromosomal abnormalities. It is also suggested that immunostaining for NPM may assist in the monitoring of minimal residual disease in a setting (normal karyotype and CD34 negativity) in which no molecular or immunophenotypic markers are available.
Furthermore these bone marrow aspirate cell blocks will be available for any future immunostaining required for the patient. We recommend also routine screening for NPMc immunostaining in all cases of primary AML in order to identify those cases expected to have a favourable prognosis and better response to induction chemotherapy.
Notes
Available online 26 August 2011
Peer review under responsibility of Alexandria University Faculty of Medicine
References
- D.G.GillilandC.T.JordanC.A.FelixThe molecular basis of leukemiaHematology (Am Soc Hematol Educ Program)20048097
- E.JaffeN.HarrisH.SteinJ.VardimanPathology and genetics of tumours of hematopoietic and lymphoid tissues2001IARC PressLyon, France
- D.GrimwadeH.WalkerF.OliverThe importance of diagnostic cytogenetics on outcome in AML: analysis of 1612 patients entered into the MRC AML 10 trial: The Medical Research Council Adult and Children's Leukaemia Working PartiesBlood92199823222333
- G.MarcucciK.MrozekC.D.BloomfieldMolecular heterogeneity and prognostic biomarkers in adults with acute myeloid leukemia and normal cytogeneticsCurr Opin Hematol1220056875
- L.BullingerK.DohnerE.BairUse of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemiaN Engl J Med350200416051616
- P.J.ValkR.G.VerhaakM.A.BeijenPrognostically useful gene-expression profiles in acute myeloid leukemiaN Engl J Med350200416171628
- C.RoumierP.FenauxM.LafageM.ImbertV.EclacheC.PreudhommeNew mechanisms of AML1 gene alteration in hematological malignanciesLeukemia172003916
- T.PabstB.U.MuellerP.ZhangDominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-alpha (C/EBPalpha), in acute myeloid leukemiaNat Genet272001263270
- S.FrohlingR.F.SchlenkI.StolzeCEBPA mutations in younger adults with acute myeloid leukemia and normal cytogenetics: prognostic relevance and analysis of cooperating mutationsJ Clin Oncol222004624633
- H.LeroyC.RoumierP.HuygheV.BiggioP.FenauxC.PreudhommeCEBPA point mutations in hematological malignanciesLeukemia192005329334
- S.SchnittgerC.SchochM.DugasAnalysis of FLT3 length mutations in 1003 patients with acute myeloid leukemia: correlation to cytogenetics, FAB subtype, and prognosis in the AMLCG study and usefulness as a marker for the detection of minimal residual diseaseBlood10020025966
- C.ThiedeC.SteudelB.MohrAnalysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosisBlood99200243264335
- A.BeghiniC.B.RipamontiR.CairoliKIT activating mutations: incidence in adult and pediatric acute myeloid leukemia, and identification of an internal tandem duplicationHaematologica892004920925
- H.KiyoiT.NaoeY.NakanoPrognostic implication of FLT3 and N-RAS gene mutations in acute myeloid leukemiaBlood93199930743080
- S.SchnittgerU.KinkelinC.SchochScreening for MLL tandem duplication in 387 unselected patients with AML identify a prognostically unfavorable subset of AMLLeukemia142000796804
- K.DohnerK.TobisR.UlrichPrognostic significance of partial tandem duplications of the MLL gene in adult patients 16 to 60 years old with acute myeloid leukemia and normal cytogenetics: a study of the Acute Myeloid Leukemia Study Group UlmJ Clin Oncol20200232543261
- C.SteudelM.WermkeM.SchaichComparative analysis of MLL partial tandem duplication and FLT3 internal tandem duplication mutations in 956 adult patients with acute myeloid leukemiaGenes Chromosomes Cancer372003237251
- B.FaliniC.MecucciE.TiacciCytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotypeN Engl J Med3522005254266
- J.L.CordellK.A.PulfordB.BigernaDetection of normal and chimeric nucleophosmin in human cellsBlood931999632642
- J.S.AndersenY.W.LamA.K.LeungNucleolar proteome dynamicsNature43320057783
- Y.W.LamL.Trinkle-MulcahyA.I.LamondThe nucleolusJ Cell Sci118200513351337
- D.L.SpectorR.L.OchsH.BuschSilver staining, immunofluorescence, and immunoelectron microscopic localization of nucleolar phosphoproteins B23 and C23Chromosoma901984139148
- R.A.BorerC.F.LehnerH.M.EppenbergerE.A.NiggMajor nucleolar proteins shuttle between nucleus and cytoplasmCell561989379390
- J.P.YunE.C.ChewC.T.LiewNucleophosmin/B23 is a proliferate shuttle protein associated with nuclear matrixJ Cell Biochem90200311401148
- Y.YuL.B.MaggiJr.S.N.BradyNucleophosmin is essential for ribosomal protein L5 nuclear exportMol Cell Biol26200637983809
- W.WangA.BudhuM.ForguesX.W.WangTemporal and spatial control of nucleophosmin by the Ran-Crm1 complex in centrosome duplicationNat Cell Biol72005823830
- J.H.ChangM.O.OlsonStructure of the gene for rat nucleolar protein B23J Biol Chem26519901822718233
- D.WangH.UmekawaM.O.OlsonExpression and subcellular locations of two forms of nucleolar protein B23 in rat tissues and cellsCell Mol Biol Res3919933342
- D.GrimwadeNPM1 mutation in AML: who and why?Blood10820063965
- B.FaliniI.NicolettiM.F.MartelliC.MecucciAcute myeloid leukemia carrying cytoplasmic/mutated nucleophosmin (NPMc+ AML): biologic and clinical featuresBlood1092007874885
- RachelRauPatrickBrownNucleophosmin (NPM1) mutations in adult and childhood acute myeloid leukemia: towards definition of a new leukemia entityHematol Oncol2742009171181
- E.AmmatunaN.I.NogueraD.ZangrilliRapid detection of nucleophosmin (NPM1) mutations in acute myeloid leukemia by denaturing HPLCClin Chem51200521652167
- G.RotiR.RosatiR.BonassoDenaturing high-performance liquid chromatography: a valid approach for identifying NPM1 mutations in acute myeloid leukemiaJ Mol Diagn82006254259
- N.I.NogueraE.AmmatunaD.ZangrilliSimultaneous detection of NPM1 and FLT3-ITD mutations by capillary electrophoresis in acute myeloid leukemiaLeukemia19200514791482
- L.I.LinT.C.LinW.C.ChouJ.L.TangD.T.LinH.F.TienA novel fluorescence-based multiplex PCR assay for rapid simultaneous detection of CEBPA mutations and NPM mutations in patients with acute myeloid leukemiasLeukemia20200618991903
- B.FaliniM.P.MartelliN.BolliImmunohistochemistry predicts nucleophosmin (NPM) mutations in acute myeloid leukemiaBlood108200619992005
- N.BolliS.GalimbertiM.P.MartelliCytoplasmic nucleophosmin in myeloid sarcoma occurring 20 years after diagnosis of acute myeloid leukaemiaLancet Oncol72006350352
- B.J.BainS.M.LewisPreparation and staining methods for blood and bone marrow filmsS.LewisB.BainI.Bates10th ed.Practical hematologyvol. 42006Churchill Livingstone6076
- I.BatesBone marrow biopsyS.LewisB.BainI.Bates10th ed.Practical hematologyvol. 62006Churchill Livingstone116130
- D.SwirskyB.J.BainErythrocyte and leukocyte cytochemistryS.LewisB.BainI.Bates10th ed.Practical hematologyvol. 132006Churchill Livingstone311334
- P.H.WiernickDiagnosis and treatment of adult acute myelocytic leukemiaP.H.WiernickG.P.CanellosJ.P.DutcherR.A.KyleNeoplastic disease of the blood1996Churchill LivingstoneNew York331351
- A.R.HandersonD.W.MossEnzymesC.A.BurtisE.R.Ashwood5th ed.Tietz fundamentals of clinical chemistryvol. 12001WB SaundersLubbock, TX352389
- D.J.NewmanC.P.PriceNonprotein nitrogen metabolitesC.A.BurtisE.R.Ashwood5th ed.Tietz fundamentals of clinical chemistryvol. 12001WB SaundersLubbock, TX414426
- K.M.GustashawChromosome stainsM.J.BarchThe ACT cytogenetics laboratory manual. The Association of Cytogenetic Technologists2nd ed.1991Raven Press Ltd.New York
- G.KingS.PayneF.WalkerG.I.MurrayA highly sensitive detection method for immunohistochemistry using biotinylated tyramineJ Pathol1831997237241
- B.FaliniI.NicolettiN.BolliM.P.MartelliA.LisoP.GorelloTranslocations and mutations involving the nucleophosmin (NPM1) gene in lymphomas and leukemiasHaematologica9242007519532
- B.FaliniN.BolliA.LisoM.P.MartelliR.MannucciS.PileriAltered nucleophosmin transport in acute myeloid leukaemia with mutated NPM1: molecular basis and clinical implicationsLeukemia2310200917311743
- J.LuoC.QiW.XuS.Kamel-ReidJ.BrandweinH.ChangCytoplasmic expression of nucleophosmin accurately predicts mutation in the nucleophosmin gene in patients with acute myeloid leukemia and normal karyotypeAm J Clin Pathol133120103440
- S.KonoplevX.HuangH.A.DrabkinCytoplasmic localization of nucleophosmin in bone marrow blasts of acute myeloid leukemia patients is not completely concordant with NPM1 mutation and is not predictive of prognosisCancer11520200947374744
- K.DohnerR.F.SchlenkM.HabdankC.SchollF.G.RuckerA.CorbaciogluMutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: interaction with other gene mutationsBlood106200537403746
- S.SchnittgerC.SchochW.KernC.MecucciC.TschulikM.F.MartelliNucleophosmin gene mutations are predictors of favorable prognosis in acute myelogenous leukemia with a normal karyotypeBlood106200537333739
- G.ZiniG.d’OnofrioEpitaph for erythroleukaemiaHaematologica8962004e82
- R.G.VerhaakC.S.GoudswaardW.van PuttenMutations in nucleophosmin (NPM1) in acute myeloid leukemia (AML): association with other gene abnormalities and previously established gene expression signatures and their favorable prognostic significanceBlood106200537473754
- M.P.MartelliV.PettirossiC.ThiedeCD34+ cells from AML with mutated NPM1 harbor cytoplasmic mutated nucleophosmin and generate leukemia in immunocompromised miceBlood11619201039073922
- D.C.TaussigJ.VargaftigF.Miraki-MoudLeukemia-initiating cells from some acute myeloid leukemia patients with mutated nucleophosmin reside in the CD34− fractionBlood11510201019761984
- T.R.KauJ.C.WayP.A.SilverNuclear transport and cancer: from mechanism to interventionNat Rev Cancer42004106117
- S.GrisendiC.MecucciB.FaliniP.P.PandolfiNucleophosmin and cancerNat Rev Cancer62006493505
- I.H.HollinkC.M.ZwaanM.ZimmermannFavorable prognostic impact of NPM1 gene mutations in childhood acute myeloid leukemia, with emphasis on cytogenetically normal AMLLeukemia232008262
- ChetsadaBoonthimatWannaThongnoppakhunChirayu U.AuewarakulNucleophosmin mutation in Southeast Asian acute myeloid leukemia: eight novel variants, FLT3 coexistence and prognostic impact of NPM1/FLT3 mutationsHaematologica9310200815651569https://doi.org/10.3324/haematol.12937