Abstract
Nosocomial spread of B. cepacia complex (Bcc) isolates amongst non-CF patients has been documented, where inadequate laboratory identification and limited treatment options are considered the main obstacles hindering accurate diagnosis and thus proper therapeutic outcome.
The present study aimed to detect the isolation percentage of Bcc from patients in Alexandria Medical University Hospital (AMUH) according to site of infection (specimen), throughout a 6 month period. Out of 2079 specimens submitted to the microbiology laboratory, 35 strains were isolated on BCSA and biochemically identified as Bcc for the first time in this laboratory.
The highest rate of isolation of Bcc isolates was from pus (85.7%) isolated from patients in the burn unit. Antibiotic susceptibility tests revealed that all Bcc isolated were Multi Drug Resistant (MDR), the highest susceptibility was to meropenem (88.5%) followed by ceftazidime (60%), tobramycin, chloramphenicol, piperacillin–tazobactam and tetracycline, while all strains were resistant to co-trimoxazole and ciprofloxacin.
Minimal Inhibitory Concentration (MIC) determining tests showed that only 11.5% were resistant to meropenem at MIC > 16 μg/ml, while 40% of the strains were resistant to ceftazidime at MIC > 32 μg/ml. Those results for the time being indicate that meropenem is the best therapeutic option for Bcc infections in AMUH.
1 Introduction
B. cepacia complex is well recognized as a significant pathogen associated with colonization and pulmonary infection in cystic fibrosis (CF) patients. However, the pathogenicity of Bcc is not always limited to CF or immunocompromised patients. Several surveys now report the increasing or simultaneous persistence of Pseudomonas aeruginosa, Acinetobacter spp. and Stenotrophomonas maltophilia nosocomial infections. As well as the emergence of newer nosocomial non-fermentative gram negative bacilli (NFGNB) such as Bcc that causes serious problems in clinical settings because of its high transmissibility between hospitalized patients and its multiple drug resistance.Citation1–Citation3 It is associated with a wide variety of infections among hospitalized patients who are usually infected via contaminated equipment or exposure to contaminated solutions. The infections usually include pneumonia, bacteremia, skin and soft tissue infection and genitourinary tract infection.Citation4–Citation6
Generally, in the literature reports of nosocomial B. cepacia infections are usually restricted to nosocomial epidemics or outbreaks. Reports of sporadic cases of B. cepacia nosocomial infections are rare, probably due to the lack of specific laboratory tests in routine testing in most hospitals, so B. cepacia has been ambiguously reported as NFGNB or simply Pseudomonas spp. This was the case in the routine microbiology laboratory in our department before this study. This also explains the lack of reports about the prevalence of B. cepacia infections in Egypt and many countries.Citation7–Citation9
In routine clinical laboratories, the identification of putative Bcc isolates is generally performed using a combination of selective media, conventional biochemical analysis and/or commercial systems. Several different media have been developed for the selective isolation of Bcc isolates from different specimensCitation10–Citation12 such as B. cepacia selective agar (BCSA), Pseudomonas cepacia (PC) agar, or oxidation-fermentation polymyxin-bacitracin-lactose (OFPBL) agar.Citation13
BCSA is more enriched than OFPBL or PCA, where yeast extract and casein provide a rich variety of ingredients that overcome the nutritional deficiencies which may prevent some strains of B. cepacia from growth on other selective media. Organisms not belonging to the Bcc that are capable of growth on BCSA include B. gladioli, Ralstonia spp. and Pandoraea spp.Citation14,Citation15
Bcc has intrinsic resistance and is one of the most antimicrobial-resistant organisms encountered. Therefore, it needs to be correctly identified and differentiated from P. aeruginosa as Bcc has inherently contrasting susceptibility pattern to that of P. aeruginosa. Bcc is intrinsically resistant to antimicrobial agents such as aminoglycosides, first-and second-generation cephalosporins, antipseudomonal penicillins and polymyxins. Thus giving extreme value to the proper identification of B. cepacia.Citation9,Citation16
The present study aimed to detect the isolation percentage of Bcc from patients in the different wards of the Alexandria Main University Hospital (AMUH) according to the site of infection (specimen) throughout a period of 6 months (June to December 2011) and to determine their antibiotic susceptibility pattern.
2 Methods
The present study was conducted on 2079 specimens submitted to the routine microbiology lab as a part of routine diagnostic services to the patients admitted in AMUH, which is a 1000 bed tertiary teaching hospital in Alexandria, Egypt, throughout a period of 6 months.
All Gram negative bacilli isolates that grew as non lactose fermenting colonies on MacConkey’s agar and were oxidase positive, were maintained as stock culture in glycerol broth and stored at −20 °C to be further tested.
The stored isolates were first subcultured on MacConkey’s agar, then picked and inoculated on BCSA (Oxoid, United Kingdom) and incubated at 37 °C for 48 to 72 h.Citation17 Reference strain of Pseudomonas aeruginosa ATCC 27853 was also cultured on BCSA as negative control strain.
All strains that grew on BCSA were further subjected to presumptive identification by the following biochemical tests:Citation18
| - | Motility (Oxoid, United Kingdom). | ||||
| - | lysine decarboxylation (Oxoid, United Kingdom). | ||||
| - | Ornithine decarboxylation (Oxoid, United Kingdom). | ||||
All isolates that were found to be ornithine decarboxylase positive, lysine decarboxylase positive and motile were selected for confirmation of their identification by RapID NF Plus system.
2.1 RapID NF plus (Remel, Lexena, Kans.)
Suspension of sufficient growth from MacConkey’s agar plate in RapID inoculation fluid (1 ml) was prepared to achieve a visual turbidity equal to #1 to #3 McFarland turbidity standard, then used to inoculate the RapID NF plus panel followed by incubation at 37 °C in the incubator for 4 h. All 10 tests were first scored before the addition of reagent providing the first test result. Then the bifunctional cavities were scored again after the addition of reagent to provide the second test result, using RapID NF Plus color guide provided by manufacturer. The results were then scored to provide a microcode, then the web based program ERIC (Electronic RapID compendium) was used for identification, based on the derived microcodes obtained from panels.
2.2 Antibiotic susceptibility testing
2.2.1 Modified Bauer Kirby disk diffusion susceptibility test
Bcc isolates were tested for their susceptibility to different antimicrobial agents and interpreted according to CLSI (2010).Citation19
Mueller–Hinton agar (Oxoid, United Kingdom) plates were inoculated with a bacterial suspension equivalent to a 0.5 McFarland Standard, then the following antibiotic susceptibility disks (Oxoid) were applied:
Ceftazidime (CAZ) 30 μg, Meropenem (MEM) 10 μg, Tetracycline (TE) 30 μg, Trimethoprim-Sulphamethoxazole (SXT) 1.25/23.75 μg, Tobramycin (TOB) 10 μg, Chloramphenicol (C) 30 μg, Piperacillin–Tazobactam (TZP) 100/10 μg, Ciprofloxacin (CIP) 30 μg. Zones of growth inhibition were measured after overnight incubation at 37oC.
2.2.2 Microbroth dilution method was used to determine the MIC for MEM and CAZ
All Bcc strains were tested for their susceptibility to Ceftazidime and Meropenem by microbroth dilution technique and using Pseudomonas aeruginosa ATCC 27853 as a control.Citation20
For each antibiotic, a stock solution was prepared at a concentration of 1000 μg/ml, then two fold serial dilution of the antibiotic was done in cation adjusted Mueller Hinton broth (CAMHB) starting from 256 μg/ml till 0.25 μg/ml in a microtitre plate as described by CLSI (2006).Citation20
Pure overnight cultures of each isolate on nutrient agar were used for the procedure. The turbidity was adjusted with sterile distilled water to 0.5 McFarland standard. Then 100 μl of the (diluted 1:100) isolate was added to an equal volume of antimicrobial solution in each well in the microtitre plate, to reach a final density = 5 × 105 cfu/ml.
Inoculated and uninoculated antibiotic-free broth as well as a complete set of antibiotic dilutions for the reference P. aeruginosa strain (ATCC 27853) were included. Then the microtitre plates were incubated at 37 °C overnight.
The MIC was recorded as the lowest concentration of antibiotic that completely inhibits the growth of the organism.
3 Results
Out of 2079 cultured specimens, 150 isolates of non fermentative gram negative bacilli (NFGNB) were found to be oxidase positive. Only 91 of these oxidase positive isolates were able to grow on BCSA. Out of these 91 strains growing on BCSA, 77 (84.6%) were lysine decarboxylase positive and 58 (63.7%) were ornithine decarboxylase positive. Thirty-five (38.5%) strains were lysine decarboxylase positive, ornithine decarboxylase positive and motile fulfilling the basic criteria for biochemical identification of Bcc which was further confirmed by RapId NF plus.
The highest percent (85.7%) of B. cepacia complex confirmed isolates by RapID NF Plus were from pus specimens, 11.4% were from sputum specimens and 2.9% were from urine specimens.
Most of strains (68.1%) were from burn unit – which were all pus specimens- followed by dermatology unit (8.7%) then chest unit (5.8%) and the remaining units showed the same isolation percent (2.9%).
Identification by RapID NF Plus:
Confirmation of identity and profiling of the Bcc isolates are shown in .
All strains were found to be multidrug resistant (resistant to three or more antimicrobial classes).
Table 1 Biochemical profile of 35 Bcc isolates identified by RapID NF Plus.
Altogether, 17 different antibiotic resistance patterns were observed as shown in . A total of 8 profiles were unique but 9 profiles were common to 2 or more isolates.
Table 2 Antibiotic resistance pattern of the 35 Bcc strains as detected by Bauer Kirby single disk diffusion technique.
It was found that 5 strains; No. 2, 8, 12, 25, 26 that shared the antibiotic resistance pattern (TE, SXT, CIP, TZP), 4 strains; No. 6, 7, 20, 22 that shared the antibiotic resistance pattern (TE, SXT, CIP, C, TZP), 3 strains; No. 1, 13, 15 that shared the antibiotic resistance pattern (CAZ, TE, SXT, CIP, TOB, C), 3 strains; No. 4, 14, 23 that shared the antibiotic resistance pattern (TE, SXT, CIP, TOB, C, TZP) and 2 strains; No. 9,11 that shared the antibiotic resistance pattern (CAZ, TE, SXT, CIP, TZP) were isolated from specimens received from the burn unit.
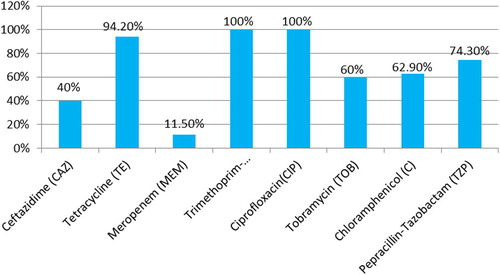
Bcc was most susceptible to ceftazidime (45.7%) and meropenem (34.5%) followed by tobramycin (22.9%), and they were found to have 100% resistance to trimethoprim-sulphamethoxazole and ciprofloxacin. Chloramphenicol had intermediate susceptibility (37.1%) (see ).
Fig. 1 Percentage of resistant strains of B. cepacia complex to different antimicrobial agents by the Bauer Kirby single disk diffusion technique.
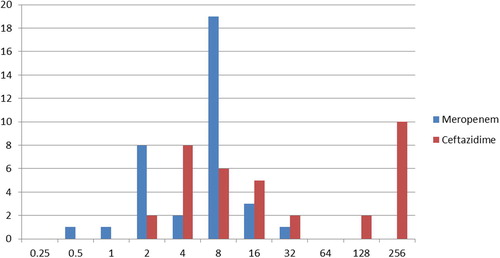
Microbroth dilution for ceftazidime showed that out of 35 strains, 16 strains were sensitive with MIC between 2–8 μg/ml, 5 strains were of intermediate sensitivity (16 μg/ml) and the remaining 14 strains were resistant with MIC ⩾ 32 μg/ml. Out of which 10 strains had MIC 256 μg/ml indicating a higher level of resistance to ceftazidime. As for meropenem, 12 strains were sensitive at MIC between 0.5–4 μg/ml, 19 strains of intermediate sensitivity 8 μg/ml and the remaining 4 strains were resistant at MIC ⩾ 16 μg/ml which indicates resistance to meropenem (see ).
4 Discussion
B. cepacia complex is now added to the list of nosocomial pathogens that cause serious problems in clinical settings because of its high transmissibility between hospitalized patients and its multiple drug resistance.Citation4–Citation6
Out of the 2079 clinical specimens received in the Microbiology Laboratory, from all hospital clinical Departments (medical wards, surgical wards and ICUs), 150/2079 (7%) isolates of NFGNB were found to be oxidase positive from which 35/150 (23%) isolates were confirmed as Bcc by RapID NF Plus. The highest rate of isolation from oxidase positive NFGNB of Bcc was from pus 30/35 (85.7%) followed by sputum 4/35 (11.4%) and urine 1/35 (2.9%).
Other studies reported higher rates of isolation of B. cepacia complex from specimens other than those in our study. Gales et al. (2005)Citation3 found that out of 176 NFGNB (83/176) belonging to Burkholderia spp.: 52/83 (62.7%) were from blood, 25/83 (30.1%) were from sputum, 3 (3.6%) were from skin and soft tissue infection and 3 (3.6%) were from urine.
On the other hand, in a Turkish University Hospital, Dizbay et al.Citation21 in 2009 showed, that 39 strains of Bcc were isolated from various clinical specimens obtained from hospitalized patients mainly in the ICU and this constituted 1.0% of gram negative isolates and was thus considered as a rare cause of nosocomial infection in this hospital.
In our study, out of 150 NFGNB oxidase positive isolates, 91 (61%) were able to grow on BCSA, 35/91 (38%) were preliminarily identified as Bcc by motility, lysine and ornithine decarboxylases and were confirmed by RapID NF Plus. The choice of BCSA in this study was based on the findings of Henry et al. in 1999Citation22 and Eram et al. in 2004.Citation17
The current study shows that most of Bcc strains isolated were from Alexandria Main University Hospital surgical wards 27/35 (77.1%), and the highest isolation among the surgical wards was from the Burn unit 24/35 (68%). This may be attributed to significant thermal injuries that induce a state of immunosuppression, in burn patients, also the devitalized tissue and moist burn are favorable conditions for colonization of micro-organisms and subsequent infection and dispersion.Citation23
The use of lysine and ornithine decarboxylation together with motility in our study for the preliminary identification of B. cepacia as well as the elimination of other oxidase positive NFGNB organisms that grew on BCSA (Ralstonia spp. and Pandoraea spp.) was based on other similar studies using biochemical tests for preliminary identification.Citation10,Citation17,Citation24 This was further confirmed by the RapID NF Plus commercial system, giving the above mentioned biochemical tests in combination with BCSA an important value in preliminary identification of B. cepacia.
A study by Shelly et al.Citation25 in 2000 identified Bcc by different commercial systems and concluded that although misidentification is widespread; the RapID NF Plus was accurate, truly rapid, easy to use and the results were easy to interpret, relatively reproducible when compared with those of the API 20NE and Vitek systems prior to supplemental testing. Because the RapID NF Plus system is enzyme based and uses carbon substrate assimilation, its ability to identify weakly oxidizing B. cepacia isolates and atypical P. aeruginosa isolates may be enhanced compared with those of the other commercial systems.
In our study, we therefore relied on the RapID NF Plus system for confirming the preliminary identification of the 35 strains of Bcc.
The most common profile was profile 2 which includes 42.8% of Bcc strains by a level of discrimination 95% followed by profile 3 which includes 31.4% of the strains by a level of discrimination 99.8% (). Fifteen strains belonging to profile 2 and 3 were found in the burn unit, concluding that the most common profiles were in burn unit.
In the current study, according to the results of antibiotic susceptibility testing by the disk diffusion method, isolates of Bcc were most susceptible to meropenem (31/35, 88.5%) followed by ceftazidime (21/35,60%), tobramycin (14/35, 40%), chloramphenicol (13/35, 37.1%), piperacillin/tazobactam (9/35, 25.7%) and tetracycline (2/35, 5.8%). All strains (35/35, 100%) were resistant to both co-trimoxazole and ciprofloxacin.
Since their recognition in 1992, several studies tested Bcc for antibiotic susceptibility.Citation7,Citation21 They all agreed that this organism was highly resistant to multiple antibiotics. Gautam et al.Citation7 in 2009 tested 30 isolated strains of Bcc and they found that their isolates were susceptible to piperacillin–tazobactam (26/30, 86.7%), levofloxacin (25/30, 83.3%), ceftazidime (24/30, 80%) and tetracycline (23/30, 76.7%). Amongst these 30 isolates; maximum resistance was against meropenem (11/30, 36.7%) and co-trimoxazole (7/30, 23.3%).
Disbay et al.Citation21 in 2009 tested 39 nosocomial strains. They reported that their B. cepacia isolates were most resistant to ceftazidime (24/39, 61.5%), followed by amikacin and ciprofloxacin (21/39, 53.8%) each, then meropenem (19/39, 48.7%), co-trimoxazole (22/39, 56.4%) and piperacillin–tazobactam (15/39, 38.4%).
Comparing the results of our study to those two studies we observe that our strains were more sensitive to meropenem (88.5%) than (51.3%) in Disbay et al. and (36.7%) in Gautam et al. but our strains were less sensitive to ceftazidime (60%) than those of Gautam et al. (80%), yet the least sensitivity to ceftazidime was reported by Disbay et al. (38.5%).
More discrepancies are observed as regards piperacillin–tazobactam, where B. cepacia complex strains in the study of Gautam et al. and Disbay et al. were more sensitive to this antibiotic than those tested in our study showing 86.7%, 61.6%, 25.7% sensitivity respectively. On the other hand none of the 35 B. cepacia complex strains in our study were sensitive to ciprofloxacin, yet 83.3% of Gautam et al. strains and 46.2% of Disbay et al. strains were sensitive to it.
The three studies agree that resistance to co-trimoxazole is common, only 23.8% of Gautam et al. strains and 43.6% of Disbay et al. strains were sensitive to it. However, our strains were 100% resistant to co-trimoxazole, although this drug was considered to be effective in treating cases of CF in earlier studies, and some cases of nosocomial infection. Those variations of antibiotic susceptibility results between different countries are probably explained by the different antibiotic policies used.Citation26,Citation27 These findings, also emphasize even more the need to isolate and test reliably more strains of Bcc to review the therapeutic measures. Determining the MIC of significant antibiotics that are still active on Bcc as tested by the modified Bauer Kirby test is therefore imperative.
In the current study, ceftazidime and meropenem were the antibiotics selected for testing their MIC on the 35 Bcc strains by the microbroth dilution method. The results of these tests conformed perfectly with the results of the modified Bauer Kirby disk diffusion technique for both antibiotics.
Gales et al. (2005)Citation3 in a large antimicrobial surveillance program between 1997 and 2002 studied the MIC of ceftazidime and meropenem as well as others isolated from Latin America. They found that susceptibility rates slightly varied from 79.5% for meropenem to 83.1% for ceftazidime. In their study ceftazidime (MIC50, 4 μg/ml; 83.1% susceptibility) was the most active β-lactam against Bcc in contrast to results reported by other studies that showed Meropenem as the most active β-lactam.Citation28,Citation29 Our study agrees with Visalli et al. and Bonacorsi et al. where if the total susceptibility of our B. cepacia complex strains is taken into account; 88.5% of our B. cepacia complex strains were inhibited by meropenem while only 60% of these strains were inhibited by ceftazidime. Apparently, even MIC results confirm that for the time being, meropenem is the best therapeutic option for Bcc infections in our Alexandria Main University Hospital.Citation29,Citation30
In the current study, the antibiotic resistance pattern of the 35 Bcc revealed that altogether the 35 strains were distributed among 17 different resistance profiles. The largest cluster of strains of identical resistance profile was from patients in the burn unit where 5 strains shared the resistance pattern TE, SXT, CIP, TZP and 4 strains shared the resistance pattern TE, SXT, CIP, TZP, C with resistance to Chloramphenicol as the only added minor variation so the 9 strains could be safely accepted to have almost identical resistance pattern which may implicate a common source or origin. In addition, 3 strains shared the antibiotic resistance pattern CAZ, TE, SXT, CIP, TOB, C and another 3 shared the pattern TE, SXT, CIP, TOB, C, TZP. This finding requires further study of the environment in the burn unit in the near future. More strains of Bcc are to be isolated and typed from both the patients and the hospital environment to detect the source of cross contamination and infection.
Conflict of interest
The authors declare no conflict of interest.
Notes
Peer review under responsibility of Alexandria University Faculty of Medicine.
Available online 18 September 2014
References
- J.R.GovanP.H.BrownJ.MaddisonEvidence for transmission of Pseudomonas cepacia by social contact in cystic fibrosisLancet34219931519
- D.A.PeguesL.A.CarsonO.C.TablanAcquisition of Pseudomonas cepacia at summer camps for patients with cystic fibrosisJ Pediatr1241994694702
- C.GalesN.JonesS.AndradeS.SaderAntimicrobial susceptibility patterns of unusual nonfermentative gram-negative bacilli isolated from Latin America: report from the SENTRY Antimicrobial Surveillance Program (1997–2002)Mem Inst Oswaldo Cruz, Rio de Janeiro10062005571578
- S.G.GefticH.HeymannF.W.AdairFourteen year survival of Pseudomonas cepacia in a salts solution preserved with benzalkonium chlorideAppl Environ Microbiol3731979505510
- R.B.JohnstonClinical aspects of chronic granulomatous diseaseCurr Opin Haematol8120011722
- A.SousaG.RamosH.LeitãoBurkholderia cepacia complex: emerging multihost pathogens equipped with a wide range of virulence factors and determinantsInt J Microbiol2011201019
- C.MukhopadhyayA.BhargavaA.AyyagariTwo novel clinical presentations of Burkholderia cepacia infectionJ Clin Microbiol42200439043905
- V.GautamP.RayG.D.PuriK.SharmaP.VandammeS.K.MadhupInvestigation of Burkholderia cepacia complex in septicaemic patients in a Tertiary Care Hospital, IndiaNepal Med Coll J1142009222224
- V.GautamL.SinghalP.RayBurkholderia cepacia complex: beyond Pseudomonas and acinetobacterIndian J Med Microbiol292011412
- J.J.LiPumaT.CoenyeP.VandammeR.W.JohnTaxonomy and identification of the Burkholderia cepacia complexJ Clin Microbiol391020013427
- D.B.ShellyT.SpilkerE.J.GracelyT.CoenyeP.VandammeJ.J.LiPumaUtility of commercial systems for identification of Burkholderia cepacia complex from cystic fibrosis sputum cultureJ Clin Microbiol38200031123115
- C.Van-PeltC.M.VerduinW.H.GoessensM.C.VosB.TümmlerC.SegondsIdentification of Burkholderia spp in the clinical microbiology laboratory: comparison of conventional and molecular methodsJ Clin Microbiol37199921582164
- T.CoenyeP.VandammeMolecular microbiology and genomics bookfirst edition.2007Taylor & FrancisUnited Kingdom
- A.BevivinoC.DalmastriS.TabacchioniL.ChiariniM.L.BelliS.PianaBurkholderia cepacia complex bacteria from clinical and environmental sources in Italy: genomovar status and distribution of traits related to virulence and transmissibilityJ Clin Microbiol402002846851
- K.VermisP.A.VandammeH.J.NelisBurkholderia cepacia complex genomovars: utilization of carbon sources, susceptibility to antimicrobial agents and growth on selective mediaJ Appl Microbiol95200311911199
- S.NzulaP.VandammeJ.R.GovanInfluence of taxonomic status on the in vitro antimicrobial susceptibility of the Burkholderia cepacia complexJ Antimicrob Chemother502002265269
- Eram S, Nejad Q, Khatami G, Nafissi N. Detection of Burkholderia cepacia complex in patients with cystic fibrosis. Tanaffos 2004; 3(9): 47–52.
- J.F.MacFaddinBiochemical tests for identification of medical bacteria3rd ed.2000Lippincott Williams and WilkinsPhiladelphiap.411–80
- Clinical and laboratory standards institute. In: Cockerill R, Wikler A, Bush K, Dudley N, Eliopoulos M, Hardy J et al., editors. Performance standards for antimicrobial susceptibility testing; 20th informational supplement. Wayne, Pennsylvania; 2010. p. 30–58.
- Cockerill R, Wikler A, Dudley N, Eliopoulos M, Craig A, Hecht W et al, editors. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, approved standard, 7th edition. Wayne, Pennsylvania: Clinical and Laboratory Standards Institute; 2006. p. 14–8.
- M.DizbayO.TunccanB.SezerF.AktasD.ArmanNosocomial Burkholderia cepacia infections in a Turkish University Hospital: a five-year surveillanceJ Infect Dev Countries342009273277
- D.HenryM.CampbellC.McGimpseyA.ClarkeL.LoudenJ.L.BurnsComparison of isolation media for recovery of Burkholderia cepacia complex from respiratory secretions of patients with cystic fibrosisJ Clin Microbiol37199910041007
- Z.NaqviQ.AzizS.KharalBurn patients effectiveness of β lactam antimicrobial drugs against gram negative bacteriaProfessional Med J1822011300305
- V.GautamP.RayP.VandammeS.S.ChatterjeeA.DasIdentification of lysine positive non fermenting gram negative bacilli (Stenotrophomonas maltophilia and Burkholderia cepacia complex)Indian J Med Microbiol2722009128133
- M.SharmaImproved cultural detection of Burkholderia cepacia from sputum in patients with cystic fibrosisJ Clin Pathol54102001803805
- D.B.ShellyT.SpilkerE.J.GracelyT.CoenyeP.VandammeJ.J.LiPumaUtility of commercial systems for identification of Burkholderia cepacia complex from cystic fibrosis sputum cultureJ Clin Microbiol38200031123115
- J.ZhouY.ChenS.TabibiL.AlbaE.GarberL.SaimanAntimicrobial susceptibility and synergy studies of Burkholderia cepacia complex isolated from patients with cystic fibrosisAntimicrob Agents Chemother51200710851088
- S.AvgeriD.MatthaiouG.DimopoulosA.GrammatikosM.FalagasTherapeutic options for Burkholderia cepacia infections beyond co-trimoxazole: a systematic review of the clinical evidenceInt J Antimicrob Agents332009394404
- M.VisalliA.S.BajaksouzianM.R.JacobsP.C.AppelbaumComparative activity of trovafloxacin, alone and in combination with other agents, against gram-negative nonfermentative rodsAntimicrob Agents Chemother41199714751478
- S.BonacorsiF.FitoussiS.LhopitalE.BingenComparative in vitro activities of meropenem, imipenem, temocillin, piperacillin, and ceftazidime in combination with tobramycin, rifampin or ciprofloxacin against Burkholderia cepacia isolates from patients with cystic fibrosisAntimicrob Agents Chemother431999213217