Abstract
Introduction
Microbial diseases are increasing year by year and they are becoming a big threat to public health. There are more than 200 known diseases transmitted by bacteria, fungi, viruses, prions, rickettsia and other microbes to humans. The emergence of drug resistance to chemical drugs is the biggest threat in controlling human pathogens. Hence novel antimicrobial agents from actinomycetes are timely needed for the control of several human pathogens.
Aim
The aim was to find some actinomycetes with antimicrobial metabolites.
Methods
Soil samples were collected from Nilgiris district in Western Ghats of Tamil Nadu, India. Actinomycetes were isolated using serial dilution and plating techniques on actinomycetes isolation agar. Streptomycin and ketoconazole (25 μg/disc) were used as reference controls. The active strains were identified by 16S rRNA and phylogenetic tree was constructed; the sequences were submitted in the GenBank.
Results
Totally 106 actinomycete strains were isolated and cross streaked against various human microbial pathogens. Only 44 (41.50%) exhibited good antimicrobial activity against different pathogenic microbes. Five isolates (FMS-20, TGH-30, TGH-31, TGH-31-1 and IS-4) were chosen for secondary screening using filtrate. Among them FMS-20 filtrate showed good inhibition on the 16th day against all tested microbial pathogens. Further the intracellular methanol extract of FMS-20 showed maximum zone of inhibition against A. brasiliensis (22 mm) at 5 mg/disc. Similarly the extracellular ethyl acetate extract of FMS-20 showed maximum zone of inhibition against B. subtilis (25 mm).
Conclusions
The present work revealed that, among 106 actinomycetes screened, Streptomyces rimosus (FMS-20) (Accession No-KT827106) showed promising antimicrobial activity against all the tested human microbial pathogens.
1 Introduction
Microbial diseases are increasing year by year and they are becoming a big threat to public health.Citation1–Citation4 There are more than 200 known diseases transmitted by bacteria, fungi, viruses, prions, rickettsia and other microbes to humans.Citation5,Citation6 Among the different microbial pathogens, viruses or prions cause 37–44% of diseases, bacteria or rickettsia cause 10–30% of diseases, protozoa cause 10.7% of diseases, fungi cause 6.3% of diseases and helminths cause 3.3% of diseases, leading to millions of death every year.Citation7–Citation9 Many bacteria excrete through faeces, which can cause undesirable effects in health and environment.Citation10–Citation13 The emergence of drug and multidrug-resistant pathogen is the biggest threat; consequently, novel antimicrobial agents from natural sources with novel mechanisms of action, are urgently needed in medical and pharmaceutical sectors.
Many research works have been carried out to control the pathogens and to identify new antimicrobial agents.Citation14–Citation16 Microbes from soils are the most important natural sources exhibiting strong biological activity against a wide range of pathogens.Citation17 Generally microbes produce bioactive molecules which are unnecessary for their growth and development but useful in defence mechanism.Citation18 Soil microorganisms in particular are intensively exploited.Citation19
Actinomycetes are Gram positive, filamentous bacteria with 55% of guanine and cytosine in their DNA.Citation20–Citation23 Actinomycetes represent one of the most important classes of bacteria for their ability to produce a wide range of biologically active secondary metabolites, which are very effective against microbial pathogens.Citation20,Citation23 More than 70–80% of all known antibiotics have been isolated from actinomycetes and are used in medicine and agriculture.Citation24 The genus Streptomyces is the biggest producer of antibiotics. Several microbial secondary metabolites are reported to be rich sources of therapeutic drugs.Citation25,Citation26
The present study aimed to evaluate some actinomycetes from different soils for antimicrobial activity against human pathogens.
2 Materials and methods
2.1 Isolation of actinomycetes from soil samples
Soil samples were collected from five different places in Western Ghats of Tamil Nadu India viz., Topslip greenhouse (TGH), Fishery mountain, (FMS), near dam mountain (NDM), Iduhatty (IS) and Kothagiri (KS). The soil samples were collected from 15 cm depth using sterile technique as per the method of Valan Arasu et al.Citation20 and transported to the laboratory. These soil samples were air-dried for 34 h at 45 °C, crushed, and sieved prior to use for isolation following established method.Citation27 The isolation of actinomycetes was done by standard serial dilution method. One gram of soil was suspended in 9 ml of sterile double-distilled water. The dilution was carried out up to 10−5 dilutions. Aliquots (0.1 ml) of 10−2, 10−3, 10−4, and 10−5 were spread on the actinomycetes isolation agar (AIA) medium containing nalidixic acid (100 mg/l) and ketoconazole (30 mg/l) and incubated at 30 °C for 7–10 days.Citation20,Citation28–Citation31 Based on the colony morphological characterization, the actinomycetes were selected and purified on ISP-2 (International Streptomyces project medium No. 2).
2.2 Morphological characterization of isolates
Morphological features of active isolates were observed with a light microscope.Citation18,Citation20,Citation30,Citation32–Citation34 Morphological features were observed in various media such as Actinomycetes isolation agar (AIA), Starch casein agar (SCA), Yeast peptone glucose agar (YPG), Bennet medium (BENNET), M3 medium (M3), Modified nutrient glucose agar medium (MNGA), ISP-International Streptomyces Project No. 2 (ISP-2), ISP-International Streptomyces Project No. 4 (ISP-4), ISP-International Streptomyces Project No. 6 (ISP-6), and ISP-International Streptomyces Project No. 7 (ISP-7). The results were recorded after incubation at 30 °C for 7–10 days.
2.3 Preliminary screening for antimicrobial activity
The antimicrobial activities of isolated actinomycetes were performed by cross streak method.Citation35 AIA plates were prepared and inoculated with isolates by single streak in the centre of petri plate and incubated at 30 °C for 10 days. The plates were then inoculated with the test organism by a single streak at 90° angles to the actinomycetes strain and incubated at 37 °C overnight. Then the antagonism of test organism was recorded. The human bacterial pathogenic organisms such as Staphylococcus aureus (MTCC-96), Micrococcus luteus (MTCC-106), Enterococcus faecalis (MTCC-439), Bacillus subtilis (MTCC-441), Staphylococcus epidermidis (MTCC-3615), Klebsiella pneumonia (MTCC-109), Enterobacter aerogenes (MTCC-111), Vibrio parahaemolyticus (MTCC-451), Yersinia enterocolitica (MTCC-840), Saccharomyces cerevisiae (MTCC-251), Shigella flexneri (MTCC-1457), Proteus vulgaris (MTCC-1771), Pseudomonas mendocina (MTCC-11808) and human pathogenic candidal strains like Candida albicans (MTCC-4748), Candida krusei (MTCC-9215), Candida tropicalis (MTCC-4370) and Candida parapsilosis (MTCC-1965) and human pathogenic fungal strains like Trichophyton mentagrophytes (MTCC-8476), Scopulariopsis sp. (MTCC-3553), Aspergillus niger (MTCC-10180), Botyritis cinerea (MTCC-2880), Epidermo floccosum (MTCC-613), Aspergillus tubingenis (MTCC-961) and Aspergillus brasiliensis (MTCC-1344) were used in the present study.
2.4 Medium optimization
The active isolates were grown in different media for the production of bioactive compounds in an orbital shaker (150 r/min) at 30 °C (M3 medium, MNGA, YPG, BENNET and ISP-2). After the growth the culture broth was centrifuged and the supernatant was used for secondary screening.
2.5 Secondary screening for antimicrobial activity
The antimicrobial activity of selected strains was tested using filtrates against all the abovementioned human microbial pathogens using Nathans agar well diffusion method.Citation36
2.6 Day optimization
For day optimization, the active strain was cultured in the selected (well grown) production media ISP-2 for up to 3–16 days following established method.Citation21,Citation37
2.7 Biochemical characterization
The active isolates were selected for biochemical studies. Biochemical characterization was done by using biochemical kit (KB014 HiAcinetobacter Identification Kit) according to the manufacturer’s protocol. For biochemical tests urea hydrolysis, acid production indole, Voges-Proskauer, citrate, lysine, ornithine, arginine, nitrate, malonate, urease, Phenylalanine-deamination, H2S production, ONPG, glucose, mannitol, xylose, inositol, sorbitol, rhamnose, sucrose, lactose, arabinose, adonitol and raffinose were carried out.
2.8 Crude extract preparation
2.8.1 Extra cellular
The total culture filtrate (20 l) of selected strain (FMS-20) was used for solvent extractions using ethyl acetate. The ratio 1:1 (v/v) of the solvent and filtrate was shaken and mixed well. The upper layer was collected using separating funnel and solvents were evaporated using rotary evaporator.
2.8.2 Intra cellular
The mycelia of the active actinomycetes were soaked in methanol and kept in shaker for about two days at 180 r/min. Finally it was filtered with blotting paper and crude extract was separated using rotary evaporator.
2.9 Antimicrobial activity of crude extracts
Antimicrobial activity of crude extract was carried out using disc diffusion method.Citation38 Petri plates were prepared with 20 ml of sterile Muller Hinton agar (MHA, Himedia) for bacteria and potato dextrose agar (PDA Himedia) for fungal pathogens. The selected human pathogens were swabbed on top of the solidified media. The crude extracts were tested at 1.25 mg, 2.50 mg and 5.0 mg/disc concentrations. Control was maintained separately; streptomycin for bacteria and ketoconazole for fungi (25 μg/disc) were used as positive controls. The loaded discs were placed on the surface of the medium and left for 30 min at room temperature for diffusion. The plates were incubated overnight at 37 °C and zones of inhibition were recorded.Citation18
2.10 Genomic DNA isolation
The genomic DNA of active strain (FMS-20) was isolated using the HipurA Streptomyces DNA purification kit-MB 527–50 pr (Himedia), according to the manufacturer’s instructions. Briefly, the pure cultures were pelleted by centrifuging for 2 min at 12,000 r/min to obtain 10–15 mg (wet weight). The cells were resuspended thoroughly in 300 μl of lysis solution. 20 μl of RNase A solution was added, mixed and incubated for 2 min at room temperature. Then 20 μl of proteinase K solution (20 mg/ml) was added. The samples were mixed and resuspended. The cells were transferred to the Hibead Tube and incubated for 30 min at 55 °C. The mixture was vortexed for 5–7 min and incubated for 10 min at 95 °C followed by pulse vortexing. Supernatant was collected by centrifuging the tube at 10,000 r/min for 1 min at room temperature. About 200 μl of lysis solution was added, mixed thoroughly by vortexing and incubated at 55 °C for 10 min. To the lysate 200 μl of ethanol (96–100%) was added and mixed thoroughly by vortexing for 15 s. The lysate was transferred to new spin column and centrifuged at 10,000 r/min for 1 min and the supernatant was discarded. The lysate was then washed in 500 μl of prewash solution which was added to the spin column and centrifuged at 10,000 r/min for 1 min and the supernatant was discarded. The lysate was then washed in 500 μl of wash solution and centrifuged at 1000 r/min for 3 min. Two hundred microlitre of the elution buffer was pipetted out and added directly into the column without spilling, and incubated for one min at room temperature. Finally the DNA was eluted by centrifuging the column at 10,000 r/min for one min.
2.11 Analysis of 16S rRNA
The primers 27F (5′AGAGTTTGATCMTGGCTCAG3′) and 1492R (5′TACGGYTACCTTGTTACGACTT 3′) were used to amplify 16S ribosomal sequence from genomic DNA in thermal cycler (ep gradient Eppendorf). The cyclic conditions are as follows: initial denaturation at 94 °C for 3 min, 35 cycles of denaturation 94 °C for 1 min, annealing 54 °C for 1 min, extension 72 °C for 2 min, and final extension 72 °C for 7 min and finally hold at 4 °C. The PCR products were confirmed by 1% agarose gel electrophoresis.Citation18,Citation20,Citation39
2.12 DNA sequence determination
Automated sequencing was carried out according to the dideoxy chain–termination method using applied Biosystems automated sequencer by Synergy Scientific Services.
2.13 Phylogenetic studies and species identification
The sequences were compared for similarity with the reference species of bacteria contained in genomic database, using the NCBI BLAST tool (http://www.ncbi.nlm.nih.gov/BLAST). The DNA sequences were aligned and phylogenetic tree was constructed based on bootstrap test of phylogeny with neighbour-joining method using MEGA4 software. The 16S rRNA sequence was submitted to the GenBank, NCBI, USA.Citation40
2.14 16S rRNA secondary structure and restriction sites analysis
The 16S rRNA secondary structure of the DNA sequence was predicted using RNA structure version 5.7 software (Mathews Lab, University of Rochester Medical Centre) and restriction site was identified using NEB cutter online tool version 2.0 (nc2.neb.com/nebcutter2/).Citation41,Citation42
3 Results
3.1 Isolation and selection of actinomycetes
The isolated strains were identified morphologically based on the colony, sporulation and pigment in the dilution plate. A total of 106 actinomycetes strains was selected and inoculated into the AIA medium for purification. The pure strains were named as FMS-20 to FMS-35, TGH-20 to TGH-35, NDM-20 to NDM-59, IS-1 to IS-8, and KS-1 to KS-26. These isolates were maintained in ISP2 medium.
3.2 Preliminary and secondary screening
All the 106 isolates were screened against human bacterial, fungal, and candida pathogens. Only 44 (41.50%) isolates showed good antimicrobial activity against pathogens. The percentage of activity to each pathogen is listed below: MTCC 96-19.8%, MTCC 106-20.7%, MTCC 439-17.9%, MTCC 441-10.3%, MTCC 3615-8.4%, MTCC MTCC 109-4.7%, MTCC 111-6.6%, MTCC 451-23.4%, MTCC 840-12.2%, MTCC 251-10.3%, MTCC 1457-22.6%, MTCC 1771-25.4%, MTCC 11808-10.3% MTCC 9215-15%, MTCC 4370-14.1%, MTCC 1965-14.1%, MTCC 4748-14.1%, MTCC 8476-14.1%, MTCC 3553-14.1%, MTCC 2030-14.1%, MTCC 2880-14.1%, MTCC 613-14.1%, MTCC 1344-14.1%, and MTCC 961-14.1%. Among them, 5 strains (TGH-30, TGH-31, TGH-31-1, FMS-20, and IS-4) showed very good activity against different human pathogens. These cultures were chosen for secondary screening ( and ). The results revealed that FMS-20 alone showed highest antimicrobial activity against all the pathogens.
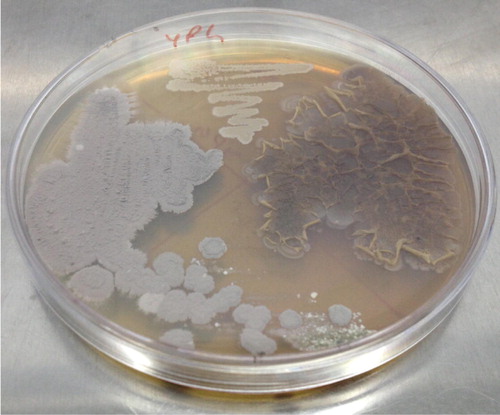
Figure 1 Preliminary screening of active isolates of Streptomyces rimosus (FMS-20) using cross streak method against different bacterial pathogens.
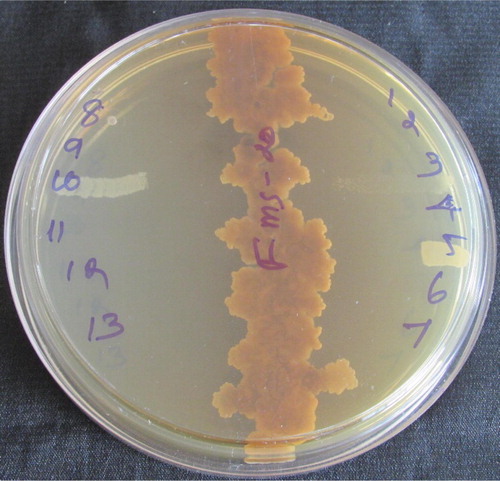
Table 1 Preliminary screening of active isolates of actinomycetes using cross streak method against different microbial pathogens.
3.3 Medium and day optimization
Among the five active strains (FMS-20, TGH-30, TGH-31, TGH-31-1, and IS-4), only FMS-20 which grew in ISP-2 medium inhibited highly all the human pathogens. The day optimization results showed that FMS-20 recorded highest zone of inhibition on 16th day.
3.4 Morphological characterization
The colony morphology of active strain (FMS-20) was noted with respect to colour, aerial and substrate mycelium, soluble pigment, colony margin, Gram staining, and growth of colony. The results are given in and .
Table 2 Morphological characterization of the active isolates of Streptomyces rimosus (FMS-20).
3.5 Biochemical characterization
The active strain (FMS-20) was selected for biochemical studies. The strain showed positive results for nitrate, glucose, mannitol, xylose, and inositol. The results are summarized in .
Table 3 Biochemical characterization of the active isolates of Streptomyces rimosus (FMS-20).
3.6 Antibacterial activity of crude extracts using disc diffusion method
Extra cellular ethyl acetate crude extract of active strain FMS-20 showed good activity against all the tested human pathogenic bacteria and candida. They did not show any activity against fungal pathogens ( and ). Intracellular methanol extract of FMS-20 also showed good activity against human fungal pathogens and it did not show any activity against bacterial and candidal pathogens ( and ).
Figure 3 Antimicrobial activity of extracellular ethylacetate crude extract of Streptomyces rimosus (FMS-20) against B. subtilis (MTCC-441).
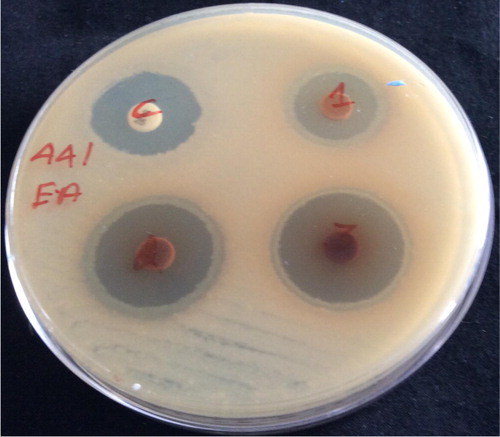
Figure 4 Antimicrobial activity of intracellular methanol crude extract of Streptomyces rimosus (FMS-20) against T. mentagrophytes (MTCC-8476).
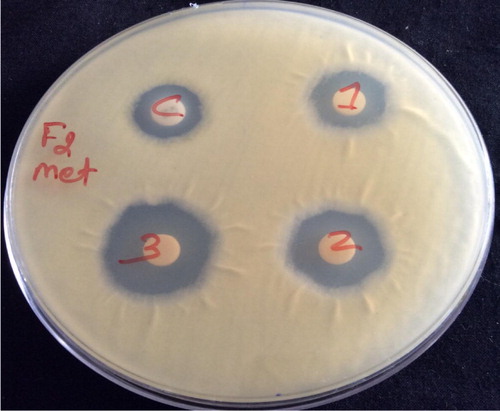
Table 4 Antimicrobial activities (in mm) of extracellular ethyl acetate crude extract of Streptomyces rimosus (FMS-20).
Table 5 Antimicrobial activities (in mm) of intracellular methanol crude extract of Streptomyces rimosus (FMS-20).
3.7 PCR amplification of FMS-20 genomic DNA
The genomic DNA was isolated using the HipurA Streptomyces DNA purification kit-MB 527-50 pr (Himedia) and the isolated genomic DNA was confirmed in 1% agarose gel stained with ethidium bromide. DNA was observed under UV Transilluminator and it showed good yield of DNA. The PCR product was analysed in 1% agarose gel electrophoresis and the size (1500 bp) was confirmed, sequenced and submitted in the GenBank (KT827106).
3.8 Molecular identification
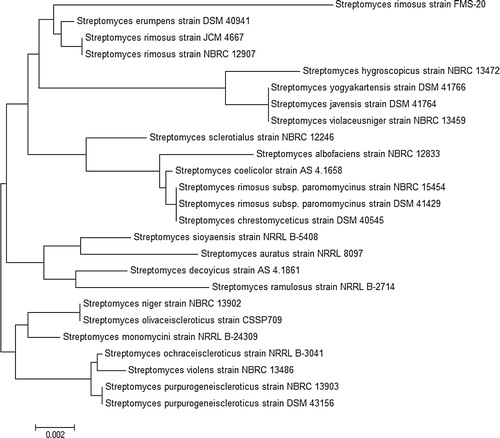
The isolated active strain FMS-20 (KT827106) showed 98% homology to Streptomyces rimosus strain NBRC 12907 16S ribosomal RNA gene partial sequence (NR_112332). The DNA sequence was aligned and phylogenetic tree was constructed by using MEGA4 software and the results revealed that FMS-20 indicated 98% similarity to S. rimosus ().
3.9 16S rRNA secondary structure and restriction sites analysis
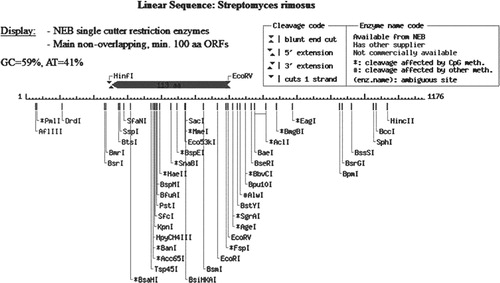
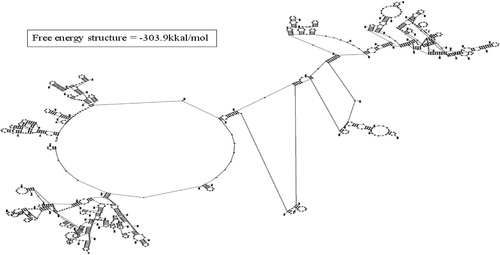
The restriction analysis of 16S rRNA sequence of FMS-20 indicated the presence of GCAT content to 59% & 41% respectively (). The energy of the predicted structure was −303.9 kkal/mol ().
4 Discussion
In the present work totally 106 actinomycete strains were isolated from five different soil samples. These strains were cross streaked against microbial pathogens. The preliminary and secondary screening results clearly showed that FMS-20 alone possessed very high antimicrobial activity against all the tested human pathogens. Oskay et al.Citation35 screened 50 actinomycetes isolated from Cyprus soil against several human pathogens. It was found that 34% of strains produced antibiotics. Valan Arasu et al.Citation43 isolated a strain of Streptomyces sp. from Western Ghats soil sample and reported that it was very effective against S. aureus, S. epidermidis, Xanthomonas sp. and C. albicans at 0.25 mg/ml concentration.
Further, in the present study, the intracellular methanol extract of FMS-20 showed maximum zone of inhibition of 22 mm against A. brasiliensis and lowest zone of inhibition of 14 mm against E. floccosum at 5 mg/disc. The extracellular ethylacetate extract of FMS-20 showed maximum zone of inhibition of 25 mm against B. subtilis and lowest zone of inhibition of 13 mm against S. flexneri at 5 mg/disc. These results were comparable with the previous report of Vijayakumar et al.Citation44 who reported that ethyl acetate extract of Streptomyces sp. was more active by producing highest zone of inhibition of 23 mm against P. vulgaris and lowest zone of inhibition of 15 mm against S. aureus. In another study Balachandran et al.Citation45 reported extra cellular ethyl acetate extract of Methylobacterium sp. with highest zone of inhibition of 13 mm against K. pneumonia and lowest zone of inhibition of 9 mm against B. subtilis. Saravanakumar et al.Citation18 reported that intracellular methanol extract of Actinobacterium showed maximum zone of inhibition of 21 mm against P. vulgaris and lowest zone inhibition of 15 mm against B. subtilis.
In several reports, ethyl acetate was mostly used as an extraction solvent to isolate the crude extracts from actinomycetes.Citation46,Citation47 Earlier several studies reported that most of the antimicrobial secondary metabolites were from extracellular actinomycetes.Citation48–Citation50 The active strain FMS-20 is taxonomically very close to S. rimosus (98% similarity). This was confirmed by blast and phylogenetic analysis. S. rimosus was Gram positive and filamentous. It showed brown colour in appearance. This strain is notably the most identified producer of oxytetracycline and other tetracycline class of antibiotics such as Polypeptides.Citation51–Citation54 Earlier, some researchers studied the crude extracts of actinomycetes on animal model. In a study, Suganthi et al.Citation55 isolated Streptomyces sp. from marine environment and checked for toxicity changes in Wistar albino rats. The extract showed no haematological, biochemical and histopathological changes in long time treatment. Similarly, Taweel et al.Citation56 tested the crude extract of actinomycetes viz., Streptomyces fradiae, Streptomyces clavus and Streptomyces roseoflavus against Ehrlich Ascites Carcinoma (EAC) on animal model using swiss albino mice. The result showed 67%, 64.2% and 75% of anticancer activity against S. fradiae, S. clavus and S. roseoflavus, respectively.
5 Conclusion
In conclusion, the extra cellular ethyl acetate crude extract of S. rimosus showed good activity against all the tested human pathogenic bacteria and candida. Similarly, intracellular methanol extract of S. rimosus also showed good activity against human fungal pathogens. The present results suggest that the isolated actinomycete S. rimosus (FMS-20) could be used as antibacterial and antifungal agent against the tested microbial pathogens.
Recommendation
The active ethyl acetate crude extract of S. rimosus may possibly be taken up further to identify the active principle and the efficacy of such active compound may also be tested with animal model.
Conflict of interest
We declare that we have no conflict of interest.
Acknowledgements
The authors are thankful to Entomology Research institute, Loyola College, Chennai for financial support and facilities. This project was partially financially supported by King Saud University, through Vice Deanship of Research Chairs.
Notes
Peer review under responsibility of Alexandria University Faculty of Medicine.
Available online 4 April 2016
References
- K.E.JonesN.G.PatelM.A.LevyA.StoreygardD.BalkJ.L.GittlemanGlobal trends in emerging infectious diseasesNature4512008990993
- D.M.MorensG.K.FolkersA.S.FauciThe challenge of emerging and reemerging Infectious diseasesNature4302004242249
- M.S.SmolinskiM.A.HamburgJ.LederbergMicrobial threats to health: emergence, detection and response2003National Academies PressWashington, DC
- S.BinderA.M.LevittJ.J.SacksJ.M.HughesEmerging infectious diseases: public health issues for the 21st centuryScience284199913111313
- D.W.AchesonFoodborne infectionsCurr Opin Gastroenterol151999538545
- G.DianeN.M.KoopmansL.VerhoefE.DuizerA.A.KaneH.S.M.OpsteeghFood-borne diseases: the challenges of 20 years ago still persist while new ones continue to emergeInt J Food Microbiol1392010315
- L.H.TaylorS.M.LathamM.E.J.WoolhouseRisk factors for human disease emergencePhilos Trans R Soc Lond3562001983989
- M.E.J.WoolhouseS.Gowtage-SequeriaHost range and emerging and reemerging pathogensEmerging Infect Dis11200518421847
- S.CleavelandM.K.LaurensonL.H.TaylorDiseases of humans and their domestic mammals: pathogen characteristics, host range and the risk of emergencePhilos Trans R Soc Lond3562001991999
- R.M.NandreA.A.ChaudhariK.MatsudaJ.H.LeeImmunogenicity of a Salmonella enteritidis mutant as vaccine candidate and its protective efficacy against salmonellosis in chickensVet Immunol Immunopathol1442011299311
- R.M.NandreK.MatsudaA.A.ChaudhariB.KimJ.H.LeeA genetically engineered derivative of Salmonella enteritidis as a novel live vaccine candidate for salmonellosis in chickensRes Vet Sci932012596603
- R.M.NandreC.V.JawaleJ.H.LeeAdjuvant effect of Escherichia coli heat labile enterotoxin B subunit against internal egg contamination in domestic fowl immunised with a live Salmonella enterica serovar enteritidis vaccineVet J19732013861867
- R.M.NandreJ.H.LeeConstruction of a recombinant-attenuated Salmonella enteritidis strain secreting Escherichia coli heat labile enterotoxin B subunit protein and its immunogenicity and protection efficacy against salmonellosis in chickensVaccine322014425431
- M.Soussi AbdallaouiN.KamalN.Guessous-IdrissiaMycoses nosocomiales systémiques à Trichosporon asahii: à proposde trois cas au CHU Ibn Rochd de Casablanca (Maroc)Rev Franc Lab41620091518
- J.HolmalahtiO.RaatikainerA.WrightH.LaatschA.SpohrO.K.J.LyngbergProduction of dihydroabikoviromycin by Strptomyces anualatus, production parameters and chemical characterization of genotoxicityJ Appl Microbiol8519986168
- R.SolankiM.KhannaR.LalBioactive compounds from marine actinomycetesIndian J Microbiol4842008410431
- K.BushM.MacielagNew approaches in the treatment of bacterial infectionsCurr Opin Chem Biol42000433439
- P.SaravanakumarJ.P.Preetjam RajV.DuraipandiyanS.IgnacimuthuAntibacterial activity of some actinomycetes from Tamil Nadu, IndiaAsian Pac J Trop Biomed22012936946
- G.H.TalbotJ.BradleyJ.E.EdwardsD.GilbertM.ScheldJ.G.BartlettBad bugs need drugs: an update of the development pipeline from the antimicrobial task force of the Infectious Diseases Society of AmericaClin Infect Dis422006657668
- M.Valan ArasuV.DuraipandiyanP.AgastianS.IgnacimuthuAntimicrobial activity of Streptomyces sp. ERI-26 recovered from Western Ghats of Tamil NaduJ Mycol Med182008147153
- A.AouicheC.BijaniA.ZitouniF.MathieuN.SabaouAntimicrobial activity of saquayamycins produced by Streptomyces spp. PAL114 isolated from a Saharan soilJ Mycol Med2420141723
- N.N.GerberH.A.LechevalierGeosmin, an earthy-smelling substance isolated from actinomycetesAppl Microbiol31965935938
- H.IkedaJ.IshikawaA.HanamotoM.ShinoseH.KikuchiT.ShibaComplete genome sequence and comparative analysis of the industrial microorganism Stretomyces avermitilisNat Biotechnol212003526531
- J.J.SanglierH.HaagT.A.HuckT.FehrReview of actinomycetes compounds: 1990–1995Expert Opin Investig Drugs51996207223
- J.D.BulockB.KristiansenBasic biotechnology1997Academic PressNew York433
- R.H.BaltzAntibiotic discovery from actinomycetes: will a renaissance follow the decline and fall?SIM News552005186196
- I.SaadounK.M.HameedA.MoussauuiCharacterization and analysis of antibiotic activity of some aquatic actinomycetesMicrobios991999173179
- C.ThawaiS.TanasupawatT.ItohK.SuwanboriruxT.KudoMicromonospora auratinigra sp. nov., isolated from a peat swamp forest in ThailandActinomycetologica182004814
- S.SinghP.KumarN.GopalanB.ShrivastavaR.C.KuhadH.S.ChaudharyIsolation and partial characterization of actinomycetes with antimicrobial activity against multidrug resistant bacteriaAsian Pac J Trop Biomed2201211471150
- E.B.ShirlingD.GottliebMethods for characterization of Streptomyces speciesInt J Syst Bacteriol161966313340
- H.S.ChaudharyJ.YadavA.R.ShrivastavaS.SinghA.K.SinghN.GopalanAntibacterial activity of actinomycetes isolated from different soil samples of Sheopur (A city of central India)Adv Pharm Technol Res422013118123
- I.L.PepperC.P.GarbaEnvironmental Microbiology2004Elsevier Academic PressSan Diego, CA3749
- S.SirvastavaV.SinghalAdvances in general microbiologyFundam Microbiol31994176177
- T.L.DeepikaK.KannibiranA morphological, biochemical, and biological studies of halophilic Streptomyces sp. isolated from saltpan environmentAm J Infect Dis532003207213
- M.OskayAntifungal and antibacterial compounds from Streptomyces strainAfr J Biotechnol813200930073017
- P.NathanE.J.LawD.F.MurphyB.G.Mac MillanA laboratory for the selection of topical antimicrobial agentsBurns41978177178
- A.PandeyA.ShuklaSkMajumdarUtilization of carbon and nitrogen sources by Streptomyces kanamyceticus M 27 for the production of an Anti-bacterial antibioticAfr J Biotechol42005909910
- A.W.BauerW.M.M.KirbyJ.C.SherriesM.TruckAntibiotic susceptibility testing by a standard single disc methodAm J Clin Pathol451966493496
- M.H.FarrisJ.B.OslonDetection of actinobacteria cultivated from environmental samples reveals bias in universal primersLett Appl Microbial452007376381
- N.SaltauM.NeiThe neighbor joining method: a new method for constructing phylogenetic treesMol Biol Evol41987406425
- L.I.BrodskyA.V.VasilievY.L.KalaidizidsY.S.OpipoyR.L.TatuoyS.I.FeranchukGenebee: the program package for biopolymer structure analysisDimacs81992127139
- L.I.BrodskyV.V.IvanovY.L.KalaidizidsA.M.LeontoyichV.K.NikolaeyS.I.FeranchuckGene bee-NET: internet-based server for analyzing biopolymers structureBiochem6081995923928
- M.Valan ArasuV.DuraipandiyanP.AgastianS.IgnacimuthuIn vitro antimicrobial activity of Streptomyces spp. ERI-3 isolated from Western Ghats rock soil (India)J Mycol Med1920092228
- R.VijayakumarK.Panneer SelvamC.MuthukumarN.ThajuddinA.PanneerselvamR.SaravanamuthuAntimicrobial potentiality of a halophilic strain of Streptomyces sp. VPTSA18 isolated from saltpan environment of Vedaranyam, IndiaAnn Microbiol62201210391047
- C.BalachandranV.DuraipandiyanS.IgnacimuthuCytotoxic (A549) and antimicrobial effects of Methylobacterium sp. isolate (ERI-135) from Nilgiris forest soil, IndiaAsian Pac J Trop Biomed292012712716
- M.M.FrancoL.E.L.CountinhoDetection of novel secondary metabolitesCrit Rev Biotechnol111991193276
- A.KavithaP.PrabhakarM.VijayalakshmiY.VenkateswarluPurification and biological evaluation of the metabolites produced by Streptomyces sp. TK-VL_333Res Microbiol1612010335345
- V.S.BernanD.A.MontenegroJ.D.KorshallaW.M.MaieseD.A.SteinbergM.GreensteinBioxalomycins new antibiotics produced by the marine Streptomyces spp. LL-31F508: taxonomy and fermentationJ Antibiot47199414171424
- H.HaceneH.DaoudiT.BhatnagarJ.C.BarattiG.LefebvreA new aminoglycosidase anti Pseudomonas antibiotic produced by a new strain of SpirillosoraMicrobiol102200069
- J.BerdyBioactive microbial metabolites: a personal viewJ Antibiot5812005126
- P.M.RhodesN.WinskillE.J.FriendM.WarrenBiochemical and genetic characterization of Streptomyces rimosus mutants impaired in oxytetracycline biosynthesisJ Gen Microbiol1241981329338
- A.Abou-ZeidN.Y.BaeshinUtilization of date-seed lipid and hydrolysate in fermentative formation of oxytetracyline by Streptomyces rimosusBioresour Technol4119924143
- N.P.ConchaB.BorovickaP.F.LongD.HranueliP.G.WatermanI.S.HunterAblation of the otcC gene encoding a post-polyketide hydroxylase from the oxytetracyline biosynthetic pathway in Streptomyces rimosus results in novel polyketides with altered chain lengthJ Biol Chem2804520053745537460
- N.SinghV.RaiOptimization of cultural parameters for antifungal and antibacterial metabolite from microbial isolate; streptomyces rimosus MTCC 10792 from soil of ChhattisgarhInt J Pharm Pharm Sci4201294101
- P.SuganthiS.RavikumarToxicity studies of crude extracts from marine Streptomyces sps. with potential antibacterial sensitivity against antibiotic resistant human pathogensAsian Pac J Trop Biomed201210701076
- F.E.TaweelM.I.A.DoubaraG.FoudaH.A.El -MezayenOncostatic effect of Streptomyces crude extracts on murine ehrlich ascites carcinomaIIJMMS182014100106