Abstract
Aim and objectives
The histone deacetylase (HDAC) inhibitor, Valproic Acid (VPA), causes growth inhibition and apoptosis in colorectal cancer cells. HDAC inhibition is associated with the transcriptional regulation of Natriuretic Peptide Receptor-A (NPR-A). NPR-A regulates voltage-gated potassium channel, KQT-like subfamily Q, member 1 (KCNQ1). NPR-A and KCNQ1 are also involved in the initiation and propagation of cancer cells. In this study, we investigated the simultaneous expressional changes of NPR-A and KCNQ1 among VPA-treated colon cancer cells.
Materials and methods
Human colorectal cancer cells were cultured and treated with increasing concentrations of VPA at different time points. MTT viability test was conducted to evaluate the growth inhibition. Real Time RT-PCR was used to quantify differential mRNA expression of NPR-A and KCNQ1 genes. Two-way ANOVA and bonferroni post-tests were used to analyze data statistically.
Results
We showed that VPA treatment inhibits the growth of SW-480 cells more efficiently compared to HT-29. NPR-A and KCNQ1 genes were significantly upregulated upon VPA treatment in both cell lines (P < 0.0001).
Conclusion
The alteration of NPR-A and KCNQ1 genes were more ordered among SW-480 cancer cells. The expressional changes of KCNQ1 and NPR-A among VPA treated human colon cancer cells follow the same pattern in similar combinations. VPA could regulate the expression of KCNQ1 through altering the mRNA expression of NPR-A.
1 Introduction
Colorectal cancer (CRC) encompasses a heterogeneous and tumor specific complex of diseases resulting from different alterations in genetic and epigenetic molecular pathways.Citation1 It is the third most common cancer and third leading cause of cancer death in the United States.Citation1 Modification of histones is an epigenetic process which is involved in gene regulation while inhibitors of Histone deacetylases (HDACs) are introduced as a novel therapeutic class of drugs against different cancer types.Citation2,Citation3 HDAC inhibitors regulate the expression of genes which are involved in specific biological responses such as apoptosis, immune regulation and angiogenesis.Citation4–Citation6 Valproic Acid (VPA) is a branched, short-chain fatty acid which acts as a HDAC1 and HDAC2a inhibitor. VPA is an established anti-epileptic drugCitation7 which is shown to induce differentiation and apoptosis in a variety of carcinoma cells.Citation8,Citation9 VPA has been also introduced as an antiproliferative compound in CRC, alone or in combination with other therapeutic strategies.Citation10
HDAC inhibitors modulate the transcriptional regulation of Natriuretic Peptide Receptor-A (NPR-A).Citation11,Citation12 NPR-A is overexpressed in numerous human cancer cells including colon adenocarcinomaCitation13–Citation17 and could be introduced as a novel anticancer target.Citation15,Citation18 Atrial Natriuretic Peptide (ANP) is the specific ligand for NPR-ACitation19 which regulates the proliferation of human gastric cancer cells via K+ voltage-gated channel subfamily Q member 1 (KCNQ1; also known as LQT1).Citation20 KCNQ1 has been also introduced as a novel regulator of cancer cell proliferation and migration in different cancer types.Citation20–Citation22 Moreover, HDAC inhibitors are reported to be capable of regulating the transcription of ATP sensitive K+ (KATP) channels,Citation23 which has not been reported among human cancer cells previously. In the present study, we investigated the effect of VPA as a HDAC inhibitor on the expression of NPR-A and KCNQ1 and also the possible simultaneous alterations of these genes in response to VPA in colon adenocarcinoma cancer cell lines.
2 Materials and methods
2.1 Cell culture and treatments
HT-29 and SW-480 human colon adenocarcinoma cells were purchased from National Cell Bank of Iran (Pasteur Institute, Iran) and cultured in RPMI 1640 (Gibco, Life Technologies, USA). HCT-116 and Caco-2 colon adenocarcinoma cells were also purchased from National Cell Bank of Iran (Pasteur Institute, Iran) and cultured in DMEM (Gibco, Life Technologies, USA). All cell culture media were supplemented with 10% FBS, 100U/ml penicillin and 100 μg/ml streptomycin. Tissue culture flasks were incubated in a fully humidified atmosphere at 37 °C with 5% CO2. 0.5 × 106 cells/mL were counted and seeded in each well of 6-well tissue culture plates followed by resuspension in complete growth media and incubated for 24 h in order to keep cells in logarithmic phase growth. HT-29 and SW-480 cells were treated with the most antiproliferative concentrations of VPA (Sigma-Aldrich, USA) (1 mM, 2 mM and 4 mM) with the least cytotoxicity effects on normal cells.Citation24 Treated and non-treated cells were then incubated for different time points (24, 48 and 72 h). Finally, the cells were detached using 0.025% trypsin-EDTA (Gibco, Life Technologies, USA) for consequent RNA extraction.
2.2 Total RNA extraction and Real Time RT-PCR
Total RNA was extracted using Biozol (Bioer, China) according to the manufacturer’s protocol. To remove any genomic DNA contamination, 1 microgram of total RNA was used for DNase I (CinnaGen, Iran) digestion and were reverse transcribed to cDNA using random hexamer primers (Bioron, Germany). Real-time RT-PCR was performed with Bioer Real-time PCR detection system (Bioer Technology, China) and Bioron SYBR green master mix (Bioron, Germany). Human 18S ribosomal RNA (18s rRNA) was used as a consistent internal control for gene expression normalization.Citation25 Gene specific primers which were designed to span exons for KCNQ1, NPR-A and 18s rRNA are summarized in . All data are presented as fold changes compared to non-treated cells.
Table1 Gene specific primers used for Real Time RT-PCR.
2.3 Cell viability (MTT) assay
The cells were counted, seeded in 96-well plate (104 cells/well) and incubated for 24 h in complete culture media. The cells were then treated with different increasing concentrations of VPA (1 mM, 2 mM and 4 mM) at different time intervals (24, 48 and 72 h). After each time point, the complete culture media was removed and a final volume of 80 μl of Phenol Red free RPMI 1640 (Gibco, Life Technologies, USA) with 20 μl of MTT solution (5 mg/mL) were added to each well and incubated for 4 h at 37 °C with 5% CO2. 100 μl DMSO was then added to each well as a cell lysis solution. Percentage of cell viability was assessed by spectrophotometry at 570 nm using ELx800 Absorbance Reader (Biotek, USA).
2.4 Statistical analysis
All of the experiments for each sample were repeated in triplicates and data were demonstrated as means ± SE (Standard Error). Statistical software SPSS22.0 and Graphpad Prism 5.04 were used for data analysis. Two-Way ANOVA with Bonferroni post hoc test was used for comparing means of multiple samples. P-values lower than 0.05 were considered as statistically significant.
3 Results
3.1 The mRNA expression level of NPR-A and KCNQ1 among different human colon cancer cell lines
The expression levels of NPR-A and KCNQ1 were quantified among four different human colon adenocarcinoma cell lines including HCT-116, SW-480, HT-29 and Caco-2. NPR-A was overexpressed in SW-480 and HT-29 cells in comparison to HCT-116 (A). The expression of NPR-A among Caco-2 cells was not statistically different compared to HCT-116. KCNQ1 was also overexpressed among HT-29 and SW-480 cells (B). Accordingly, HT-29 and SW-480 with the highest expression of NPR-A and KCNQ1 were chosen for the next phase of the study.
Fig. 1 The mRNA expression level of NPRA and KCNQ1. NPRA and KCNQ1 are overexpressed in HT-29 (A) and SW-480 (B) cell lines in comparison to other human colon cancer cells. Data are expressed relatively to mRNA levels in HCT 116 cell line, arbitrarily set at the value of 1. Data are obtained from experiments in triplicates and presented as means ± SE. Significant differences are evaluated using independent samples t-test. P-values lower than 0.05 are considered statistically significant. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns: not significant.

3.2 VPA is an effective inhibitor of proliferation in human colon cancer cells
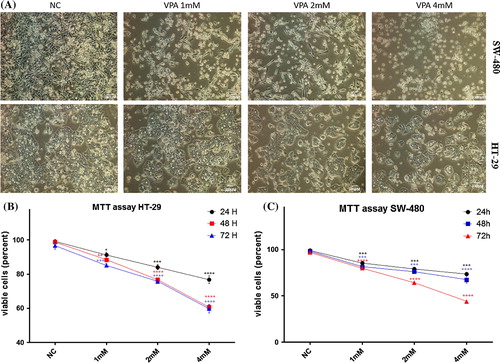
To measure the effects of VPA on cell viability, various increasing concentrations of VPA were applied to cell culture medium of SW-480 and HT-29 cancer cells at different time intervals (A). Percentages of viable cells were measured by MTT Assay. VPA treatment has reduced the viability of HT-29 (B) and SW-480 (C) cells significantly after 24, 48 and 72 h. VPA is a more effective inhibitor of proliferation on SW-480 cancer cells.
Fig. 2 Effects of VPA on cell proliferation in vitro. (A) Growth inhibition was observed upon VPA treatments (1 mM, 2 mM and 4 mM) at different time points (24 h, 48 h and 72 h) on HT-29 and SW480 cells. MTT assay was performed to determine the proliferation of HT-29 (B) and SW-480 (C) human colon cancer cells. VPA inhibited cell viability of SW480 and HT-29 cells in a concentration and time dependent manner. Data represent the mean ± SE from three independent experiments. Significant differences are evaluated using independent samples t-test. P-values lower than 0.05 are considered statistically significant. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, NC: non-treated cells (mock).
3.3 The expression pattern of NPR-A is different among HT-29 and SW-480 cell lines
During the first 24 h, NPR-A was significantly overexpressed in HT-29 cells at 1 mM concentration of VPA. After 48 h, NPR-A was downregulated by increasing the concentrations of VPA. However, no significant difference was observed. However, NPR-A was elevated gently with increasing VPA concentrations after 72 h (A).
Fig. 3 NPRA expressional alterations. The mRNA expressional changes of NPRA were quantified by Real-Time RT-PCR in each combination representing different patterns among HT-29 (A) and SW-480 (B) human colon cancer cells. Data are expressed relatively to mRNA levels in non-treated (NC) cell lines, arbitrarily set at the value of 1. Data are obtained from experiments in triplicates and presented as means ± SE. Significant differences are evaluated using independent samples t-test or one-way ANOVA followed by bonferroni post hoc test while studying multiple comparisons. P-values lower than 0.05 are considered statistically significant. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, NC: non-treated cells (mock).
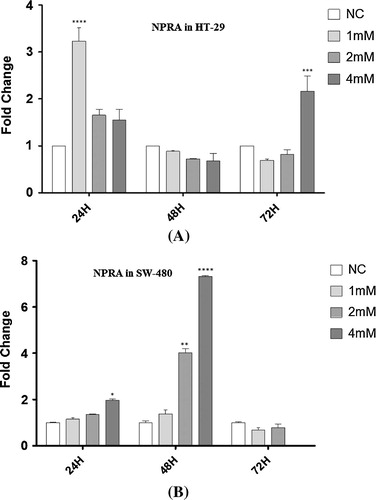
Unlike HT-29 cells, NPR-A was overexpressed among SW-480 cells in an ordered manner after 24 and 48 h. SW-480 cells treated with 4 mM VPA showed the highest expression of NPR-A. After 72 h, the expression of NPR-A decreased in all treatments and was the same as non-treated SW-480 cells (B).
3.4 KCNQ1 is overexpressed among HT-29 and SW-480 cells in response to VPA
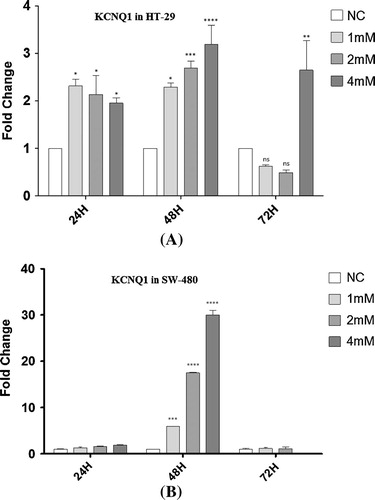
KCNQ1 was elevated gradually among HT-29 colon cancer cells proportional to the increasing concentrations of VPA after 24 and 48 h. HT-29 cancer cells showed the highest expression level of KCNQ1 at 4 mM VPA treatment after 48 h. KCNQ1 was downregulated after 72 h among VPA treated HT-29 cells except for 4 mM concentration (A). KCNQ1 increased significantly among SW-480 cells only after 48 h of incubation in all concentrations (B).
Fig. 4 Overexpression of KCNQ1 upon VPA treatment. The mRNA expression levels of KCNQ1 were quantified by Real-Time RT-PCR in each combination among HT-29 (A) and SW480 (B) human colon cancer cells. KCNQ1 is overexpressed in all VPA treated colon cancer cells. Data are expressed relatively to mRNA levels in non-treated (NC) cell lines, arbitrarily set at the value of 1. Data are obtained from experiments in triplicates and presented as means ± SE. Significant differences are evaluated using independent samples t-test or one-way ANOVA followed by bonferroni post hoc test while studying multiple comparisons. P-values lower than 0.05 are considered statistically significant. ***P < 0.001, ****P < 0.0001, NC: non-treated cells (mock).
3.5 Concurrent alterations of KCNQ1 and NPRA in colon cancer cells
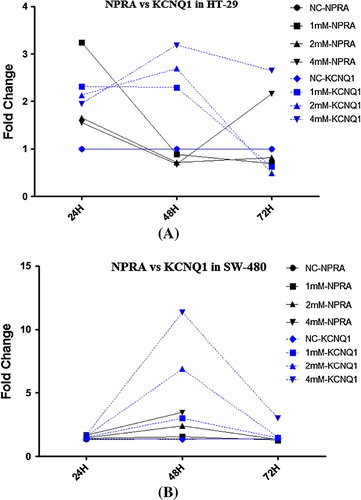
In order to estimate the probable simultaneous changes in the expression of NPR-A and KCNQ1 in human colon cancer cell lines, the expression pattern of all concentrations at each time point were adapted to its relevant. There was a correlation between the expressional changes of NPR-A and KCNQ1 among SW-480 cells. In all concentration of VPA and all tested time points, altering the expression of NPR-A directly affected the expression of KCNQ1. Such a concordance between the expression of NPR-A and KCNQ1 is observed in HT-29 cells except for treatments at 4 mM VPA ().
Fig. 5 NPRA and KCNQ1 simultaneous mRNA expression patterns. The quantified mRNA expression of NPRA and KCNQ1 genes were compared in order to derive a possible alteration pattern. We showed that the expressional changes in NPRA and KCNQ1 occur concurrently among SW-480 human colon cancer cells (B) and not HT-29 (A). Data are obtained from experiments in triplicates and presented as means. NC: non-treated cells (mock).
4 Discussion
Histone deacetylation is an epigenetic modification in gene expression which has diverse effects on gene function, malignant cellular transformation, cell cycle and apoptosis.Citation5,Citation26,Citation27 These biological and pathological mechanisms are commonly modified by HDAC inhibitors which mostly possess anticancer properties.Citation2,Citation3,Citation11 VPA is a HDAC1 and HDAC2a inhibitor which has been utilized in the therapy of epilepsy for a long time.Citation7,Citation8,Citation23 VPA alongside with other HDAC inhibitors have shown promising antiproliferative effects in different cancers including CRC.Citation28–Citation30 Moreover, the combination of VPA with conventional anticancer therapies has enhanced the effectiveness of current.Citation30,Citation31
Here, we conducted MTT viability test to measure the growth inhibitory effects of various concentrations of VPA on SW-480 and HT-29 cancer cells. The antiproliferative effects of VPA have been evaluated in different colon cancer cell lines. According to these reports, SW-480 is one of the most sensitive colon cancer cell lines to VPA treatment.Citation28 On the other hand, it has been reported that HT-29 as an APC mutant CRC cell line is less sensitive to VPA.Citation29 We showed that, VPA treatment has reduced the viability of SW-480 and HT-29 cells after each treatment. In accordance with the previous studies, we noted that VPA is a more effective proliferation inhibitor on SW-480 cancer cells compared to HT-29. However, VPA still preserves its growth inhibitory properties against HT-29 cells especially at higher concentrations.
NPR-A which is one of the specific receptors for ANP cardiac hormone is expressed in numerous human cancer cells including CRC.Citation15,Citation18 NPR-A is involved in different physiological and pathological processes and has been recently introduced as a novel anticancer target.Citation20,Citation32 However, there is no record available comparing the expression level of NPR-A among different human CRC cell lines.Citation33 In order to target the CRC cell lines with the overexpression of NPR-A, we examined the expression of NPR-A among four different human CRC cell lines including HCT-116, SW-480, HT-29 and Caco-2. The lowest expression was demonstrated in HCT-116. The expression level of NPR-A was higher among SW-480 and HT-29 cells compared to HCT-116. We also quantified the expression of KCNQ1 gene among all CRC cell lines. Similar to the expression pattern of NPR-A, the mRNA expression of KCNQ1 was higher in SW-480 and HT-29 cells. Therefore, SW-480 and HT-29, with different molecular characteristics, were selected to be studied. These CRC cell lines could be appropriate in vitro models to investigate NPR-A mediated novel anticancer strategies. The expression of NPR-A was not statistically different in Caco-2 compared to HCT-116.
HDAC inhibitors have been shown to enhance NPR-A expression by blocking HDACs and interacting with Sp1, histone acetyltransferase, and acetylated histones.Citation11 Kumar et al claimed that Trichostatin-A (TSA) as a class I HDAC inhibitor modulates the transcriptional regulation of NPR-A gene.Citation12 On the other hand, it was revealed that NPR-A knockdown could downregulate the expression of voltage-gated potassium channel, KCNQ1.Citation20 KCNQ1 channel has been introduced as a novel regulator of cancer cell proliferation in many human cancer cells including CRC. HDAC inhibitors can regulate the ATP sensitive K+(KATP) channels subunit transcription in cardiomyocytes as well,Citation23 but to date no study has explained the possible effects of HDAC inhibition on the expression of voltage-gated K+ channels including KCNQ1.
We postulated that the effects of VPA might be carried out through regulating the expression of NPR-A and consequently KCNQ1. Therefore, we aimed to investigate the anticancer effects of various concentrations of VPA at different time points on selected CRC cell lines by quantifying the alterations in the expression of NPR-A and KCNQ1 genes and the possible relation between the these two genes.
VPA might have a more transient effect on the expression of NPR-A among HT-29 CRC cancer cells. We noted that, at lower concentrations of VPA which induce the least toxicity, NPR-A is overexpressed but an unclear mechanism (maybe a negative feedback) reverses the expression pattern of NPR-A after 24 h and the expression was reduced. Unlike HT-29 cells, the transcriptional effect of VPA on the expression of NPR-A is more persistent among SW-480 cells. The expression of NPR-A is finally decreased in all treatments and reaches the same as non-treated SW480 cells. Moreover, KCNQ1 is overexpressed among HT-29 and SW-480 colon cancer cells in response to VPA. This overexpression is more vividly observed among HT-29 cells in higher concentrations of VPA at all-time points. The expression level of KCNQ1 among SW-480 cells increased only after 48 h of incubation in all concentrations. The alterations in the expression of NPR-A and KCNQ1 among SW-480 cells occur simultaneously with the same pattern. This pattern was also observed in the expression of NPR-A and KCNQ1 among HT-29 cells with minimal irregularity. Accordingly, VPA alters the expression of NPR-A and KCNQ1 with the same pattern. However, these alterations are more ordered among SW-480 human CRC cells.
5 Conclusion
VPA mediates its effects on human CRC cell lines through altering the expression of NPR-A and KCNQ1 simultaneously. These alteration patterns are more ordered among SW-480 cancer cells. We propose the alterations in the expression of NPR-A and KCNQ1 as a novel molecular mechanism of effect for the effects of VPA in CRC. Our findings need to be investigated in further experiments using more sophisticated in vitro or in vivo examinations.
Notes
Peer review under responsibility of Alexandria University Faculty of Medicine.
Available online 28 April 2017
References
- V.AranA.P.VictorinoL.C.ThulerC.G.FerreiraColorectal cancer: epidemiology, disease mechanisms and interventions to reduce onset and mortalityClin Colorectal Cancer2016
- H.-J.KimS.-C.BaeHistone deacetylase inhibitors: molecular mechanisms of action and clinical trials as anti-cancer drugsAm J Transl Res32011166179
- A.A.LaneB.A.ChabnerHistone deacetylase inhibitors in cancer therapyJ Clin Oncol27200954595468
- A.C.WestR.W.JohnstoneNew and emerging HDAC inhibitors for cancer treatmentJ Clin Invest12420143039
- K.J.FalkenbergR.W.JohnstoneHistone deacetylases and their inhibitors in cancer, neurological diseases and immune disordersNat Rev Drug Discov132014673691
- S.KaypeeD.SudarshanM.K.ShanmugamD.MukherjeeG.SethiT.K.KunduAberrant lysine acetylation in tumorigenesis: implications in the development of therapeuticsPharmacol Ther2016
- E.BoudadiH.StowerJ.A.HalsallThe histone deacetylase inhibitor sodium valproate causes limited transcriptional change in mouse embryonic stem cells but selectively overrides polycomb-mediated Hoxb silencingEpigenet Chromatin620131
- R.FujikiA.SatoM.FujitaniT.YamashitaA proapoptotic effect of valproic acid on progenitors of embryonic stem cell-derived glutamatergic neuronsCell Death Dis42013e677
- D.WangY.JingS.OuyangInhibitory effect of valproic acid on bladder cancer in combination with chemotherapeutic agents in vitro and in vivoOncol Lett6201314921498
- J.FengJ.CenJ.LiHistone deacetylase inhibitor valproic acid (VPA) promotes the epithelial mesenchymal transition of colorectal cancer cells via up regulation of SnailCell Adh Migr92015495501
- P.KumarS.TripathiK.N.PandeyHistone deacetylase inhibitors modulate the transcriptional regulation of guanylyl cyclase/natriuretic peptide receptor-A gene interactive roles of modified histones, histone acetyltransferase, p300, and Sp1J Biol Chem289201469917002
- P.KumarR.PeriyasamyS.DasS.NeerukondaI.ManiK.N.PandeyAll-trans retinoic acid and sodium butyrate enhance natriuretic peptide receptor a gene transcription: role of histone modificationMol Pharmacol852014946957
- A.SerafinoP.PierimarchiAtrial natriuretic peptide: a magic bullet for cancer therapy targeting Wnt signaling and cellular pH regulatorsCurr Med Chem2120142401
- J.ZhangZ.ZhaoJ.WangNatriuretic peptide receptor A as a novel target for cancerWorld J Surg Oncol1220141
- X.WangP.RauljiS.S.MohapatraNatriuretic peptide receptor A as a novel target for prostate cancerMol Cancer10201156
- A.A.KasparJ.M.ReichertFuture directions for peptide therapeutics developmentDrug Discov Today182013807817
- J.BelotteN.M.FletcherM.AlexisSox2 gene amplification significantly impacts overall survival in serous epithelial ovarian cancerReprod Sci19337191145420212014
- X.KongX.WangW.XuNatriuretic peptide receptor a as a novel anticancer targetJ Cancer Res682008249256
- M.KuhnMolecular physiology of natriuretic peptide signallingClin Res Cardiol9920047682
- J.ZhangZ.ZhaoC.ZuAtrial natriuretic peptide modulates the proliferation of human gastric cancer cells via KCNQ1 expressionOncol Lett62013407414
- A.GiraultA.PrivéN.T.N.TrinhIdentification of KvLQT1 K+ channels as new regulators of non-small cell lung cancer cell proliferation and migrationInt J Clin Oncol442014838848
- W.ZhanpingP.XiaoyuC.NaW.ShenglanW.BoVoltage-gated K+ channels are associated with cell proliferation and cell cycle of ovarian cancer cellGynecol Oncol1042007455460
- N.FatimaD.C.CohenG.SukumarHistone deacetylase inhibitors modulate KATP subunit transcription in HL-1 cardiomyocytes through effects on cholesterol homeostasisFront Aging Neurosci62015
- A.PapiA.FerreriF.GuerraM.OrlandiAnti-invasive effects and proapoptotic activity induction by the rexinoid IIF and valproic acid in combination on colon cancer cell linesAnticancer Res32201228552862
- C.GuoE.RosohaM.B.LowryN.BorregaardA.F.GombartCurcumin induces human cathelicidin antimicrobial peptide gene expression through a vitamin D receptor-independent pathwayJ Nutr Biochem242013754759
- B.Barneda-ZahoneroM.ParraHistone deacetylases and cancerMol Oncol62012579589
- S.SharmaT.K.KellyP.A.JonesEpigenetics in cancerCarcinogenesis3120102736
- A.Chavez-BlancoC.Perez-PlasenciaE.Perez-CardenasAntineoplastic effects of the DNA methylation inhibitor hydralazine and the histone deacetylase inhibitor valproic acid in cancer cell linesCancer Cell Int620062
- X.HuangB.GuoAdenomatous polyposis coli determines sensitivity to histone deacetylase inhibitor-induced apoptosis in colon cancer cellsCan Res66200692459251
- X.ChenP.WongE.RadanyJ.Y.WongHDAC inhibitor, valproic acid, induces p53-dependent radiosensitization of colon cancer cellsCancer Biother Radiopharm242009689699
- K.CamphausenD.CernaT.ScottEnhancement of in vitro and in vivo tumor cell radiosensitivity by valproic acidInt J Cancer1142005380386
- E.AbdelalimI.TooyamaNPR-A regulates self-renewal and pluripotency of embryonic stem cellsCell Death Dis22011e127
- A.SerafinoN.MoroniR.PsailaAnti-proliferative effect of atrial natriuretic peptide on colorectal cancer cells: evidence for an Akt-mediated cross-talk between NHE-1 activity and Wnt/β-catenin signalingBiochem Biophys Acta1822201210041018
