Abstract
Background: Zoonotically acquired HEV has been described as one of the most successful zoonotic viral infections in human history.
Aim: In this study we characterized HEV comparative genomic analysis as it relates to swine HEV.
Materials and methods: A total of 82 chronic hepatitis B patients were recruited from May 2015 to May 2016 for this study. We conducted a serological and molecular investigation of HEV among these patients. The detected HEV were sequenced and the genomes and deduced amino acids were characterized using molecular evolutionary genetic analysis software version 7.
Results: Of the 82 chronic hepatitis B patients that were tested, 9.8% (8/82) were found to be HEV positive. Phylogenetic analysis of the HEV RNA sequences showed they are of genotype 4 and demonstrated high sequence identity with a swine isolate from China, with variation in amino acids at position 22, where leucine was present in the Malaysian human isolate while phenylalanine was present in the China swine isolate.
Conclusion: Comparative analysis of the human HEV ORF-2 nucleotide sequence suggest zoonotic origin.
1 Introduction
Hepatitis E virus (HEV) is distributed worldwide, infecting about two billion people with 14 million symptomatic infections and an annual mortality of 300,000. Hepatitis E viral infection in developing countries is endemic, but an increasing number of sporadic as well as authochronous infections have been reported in recent years.Citation1 According to the Southeast Asia Regional Office of the World Trade Organization (WTO), Asia hosts 6.5 million symptomatic HEV infections with 160,000 annual mortalities and 2700 stillbirths due to HEV infection in pregnancy. As such, more than 50% of the global HEV infection mortality occurs in Asia.Citation2 Hepatitis E virus is transmitted enterically leading to self-limiting acute viral hepatitis infection with no chronic stage.Citation3 However, in some cases, fulminant hepatic failure can result, leading to increases in morbidity and mortality, especially among high risk groups such as, pregnant women immunocompromised, elderly people and pre-existing liver diseases patients such as with chronic hepatitis B or C infections.Citation4–Citation6
Zoonotic potential of HEV has been reported in a previous study in which HEV antibodies was found in animal serum.Citation7,Citation8 For instance, some HEV cases in Japan were found to be directly associated with eating raw or under cooked deer meat,Citation9 while several cases of HEV were epidemiologically linked to consumption of poorly cooked wild boar meat or pork.Citation10–Citation12 HEV was reported to be successfully transmitted from animal to human,Citation1 as it causes acute infection as well as aggravates any existing chronic hepatitis condition. Malaysia is a multicultural and multi-ethnic society, and a center for tourist attraction and business investment. It is therefore important to study the possible risk of HEV transmission in relation to possible zoonosis.
2 Materials and methods
2.1 Study population
Chronic hepatitis B patients that present with cirrhosis, non-cirrhosis or acute flare ups seen by or admitted to the Hepatology Department of Hospital Selayang who consented to the study were recruited. However, patients with non-hepatitis B liver cirrhosis or non-cirrhotic liver disease and those with HIV co-infection were excluded from the study.
2.2 Ethical consideration
Ethical approval to conduct this study was obtained from the Malaysian Medical Research Institute (NMRR-14-1136-21909) and the Malaysian Medical Research and Ethic Committee (KKM/NIHSEC/P15-63) as well as the Universiti Putra Malaysia Human Ethic Committee (UPM/TNCPI/RMC/JKEUPM/1.4.18.1/F1). Signed informed consent was obtained from the patients by the attending physician.
2.3 Sample collection
A total of 82 patients were recruited from May 2015 to May 2016. Ten millilitres of blood was collected from each patient into an EDTA blood collecting tube, and the plasma was separated immediately after collection and kept in −80 °C before performing experiment.
2.4 Serological assay
Hepatitis E virus antibodies (IgM and IgG) were determined using enzyme linked immunosorbent assay (ELISA) according to manufacturer’s instructions (Wantai Bio-Pharm., Beijing, China). Briefly, both reagents and the frozen plasma were allowed to reach room temperature. Into the microplate ELISA well, 50 µl of sample diluent was added except the blank well. Both the plasma sample and the controls were also added to the diluents and mixed by gentle tapping. The mixture was then covered and incubated for 30 min at 37 °C, followed by 45 s soaking periods and washing five times. Horse radish peroxidase (HRP) was added and incubated at 37 °C for 30 min followed by washing five times as mentioned earlier. Fifty microliters each of chromogen A and B solution was added to the entire well, including the blank and incubated for 15 min at 37 °C, followed by adding 50 µl stop solution and the absorbance was measured using TECAN sunrise (Männedorf, Switzerland) microplate absorbance reader at a reference wavelength of 630 nm.
2.5 HEV RNA isolation
Hepatitis E viral RNA was extracted from the patients’ plasma using QIAamp RNA blood mini kit as described by the manufacturers (Qiagen, Hilden, Germany). Concisely, 20 µl protease was added to the bottom of 1.5 ml tube, followed by addition of 200 µl plasma. Buffer AL was added to all of the samples and was mixed thoroughly for 15 s by pulse-vortexing after which it was incubated for 10 min at 56 °C using a water bath. After incubation, 200 µl of ethanol was added to the sample and was vortexed for 15 s. The mixture was then emptied into 2 mL QIAamp mini columns and centrifuged at 8000 rpm for 60 min, after which the follow through was discarded. Washing step was performed by adding 500 µl of washing buffer AW1 and centrifuged at 8000 rpm for 60 min, followed by AW2 buffer centrifuged at 14,000 rpm for three minutes. The RNA was then eluted by adding 50 µl of elution buffer, and the RNA recovered was kept at −70 °C. The RNA concentration was determined by the NanoDrop® ND-1000A spectrophotometer (NanoDrop Technologies Inc.). The purity of the sample was determined by using the A260/A280 ratio to determine its purity.
2.6 HEV RNA detection and sequencing
Two published primers were used for the outer and inner polymerase chain reaction (PCR). The outer primers are: HEV-R-Outer (5′CCCTTATCCTGCTGAGCATTCTC-3′) and HEV-R-Outer (5′-AAYTATGCWCAGTACCGGGTTG-3′), and the nested or inner primers are: HEV-F-Inner (5′-GTYATGYTYTGCATACATGGCT-3′) and HEV-R-Inner (5′-AGCCGACGAAATYAATTCTGTC-3′). The primers were commercially synthesised (IDT) and were used to amplify the targeted HEV ORF-2 region.Citation13
The RT-PCR was performed using Access kit according to manufacturer’s instruction (Promega, USA). The reaction mixture was prepared in a 50 µl thin wall tube, and the reverse transcription was performed at 50 °C for 30 min, followed by 35 cycles of amplification at 94 °C for 1 min, 50 °C for 45 s, and 72 °C for 1minute, with a final extension of 72 °C for 10 min. The amplification product of the RT-PCR was used as template for nested PCR using the inner primers. Both the reaction mixture volume and the PCR conditions used for the nested PCR are the same as that of the first round described above. The amplified nested PCR products were analysed using gel electrophoresis and visualised using gel Doc. (BioRAD), with a final amplification product of 345 bp.
2.7 Sequence analysis
The partial HEV ORF2 region from the eight Malaysian HEV strains was subjected to a nucleotide BLAST search for sequence homology. The phylogenetic study was done by comparing the Malaysian HEV sequence with reference isolates from humans, pigs and wild boars retrieved from national center for biotechnology information (NCBI) gene data base. All eight sequences were submitted to the gene bank and were assigned accession numbers from KX426575 to KX426582. The maximum likelihood (ML) phylogenetic method was performed with the aid of MEGA7 softwareCitation14 for genotyping and sequence relatedness with reference human and animal strains of HEV from different sources. Model with the lowest Bayesian criterion information explaining substitution pattern among nucleotide sequences is considered the best modelCitation15 and was considered during sequence analysis. The reliability of the phylogenetic tree was determined using bootstrappingCitation16 Evolutionary divergence between sequences was estimated using the ML model. Amino acids deduced from nucleotide sequences of our isolate and that of a reference isolate from swine hepatitis were aligned using the Biological Sequence Alignment Editor, v 7.2.0 (BioEdit)Citation17 bioinformatics software to determine the amino acid differences between the human and swine HEV isolates.
2.8 Data analysis
Statistical analysis to determine the proportion of HEV infection among the study population and their demographic profile was done using statistical package for social science version 22.0 (SPSS, Inc., Chicago). Sequence analysis and phylogeny was performed using Molecular Evolutionary Genetic Analysis (MEGA5) software.Citation14
3 Results
Of the 82 chronic hepatitis B patients that were tested for antibodies against HEV, 59.8% (49/82) were male. The distribution according to ethnicity was 32.9% (27/82) Malay, 62.2% (51/82) Chinese, 3.7% (3/82) Indian and 1.2% (1/82) Orang Asli. Half (41/82) of the patients were older than 50 year of age and only 11% (9/82) were below 30 years of age
the remaining 39% (32/82) of patients were between 30 and 49 years of age. 9.8% (8/82) were found to be positive. Most of the HEV-seropositive patients were 61 years old and older (75%), followed by those within the age group of 51–60 years. Out of the eight hepatitis E infected patients, five were males (62.5%) and three females (37.5%), and all of them were Chinese. Hepatitis E virus RT-PCR was positive for all eight seropositive samples, and sequencing of the amplified PCR product confirmed the genome to be HEV. Phylogenetic analysis of this sequence confirmed that it was genotype 4 () based on evolutionary history inferred by maximum likelihood according to the Hasegawa-Kishino model.
Fig. 1 Hepatitis E virus genotyping by molecular phylogenetic analysis. The evolutionary relationship was inferred using maximum likelihood based on the model by Hasegawa-Kishino-Yano.Citation18 The tree has the highest likelihood (−2728.90830) and superior log likelihood and topology. A distinct Gamma distribution was used in evolutionary modelling (+G, parameter = 0.2081). The tree is drawn to scale 0.1, with the branch length measured in the number of substitutions per site. The red dots indicate Malaysian isolates belonging to genotype 4. G1, G2, G3, and G4 indicate genotype 1, 2, 3 and 4 respectively.
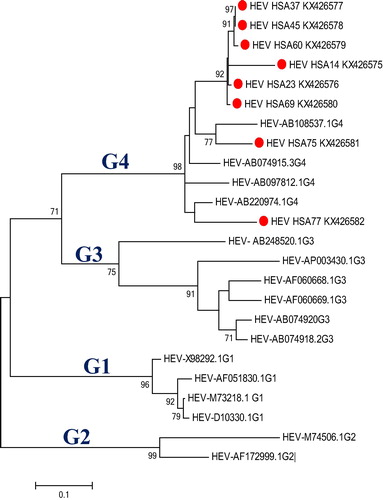
The HEV ORF-2 nucleotide sequence homology between Malaysian sequences and that of genotypes 1–4 were 78%, 75%, 80% and 96%, respectively. Homology of the Malaysian sequences in terms of geographical relatedness to other Southeast Asian countries revealed most of the Malaysian isolates are closely related to reference isolates from Laos, India and Thailand among others. However, one of the Malaysian isolates, HSA14, differed from the others and is related to isolates from Japan, Nepal and China. Phylogenetic analysis revealed that the Malaysian HEV sequences are placed in two distinct clusters with the majority (7/8) grouped in a single cluster, and they are phylogenetically related to isolates from Thailand, Laos and India. Furthermore, the remaining one Malaysian isolate was grouped with other reference isolates in a different cluster ().
Fig. 2 Molecular phylogenetic analysis of 63 nucleotide sequences which involved the first, second, third, and noncoding codon positions for a total of 4504 positions. Geographical relatedness of our Malaysian sequences (red dot), with representative sequences of Southeast Asia Countries, separate into two distinct clusters (Red and Blue). The Maximum likelihood molecular phylogenetic analysis method was used to infer the evolutionary history of this isolate based on the model by Hasegawa-Kishino-YanoCitation18 with discrete Gamma distribution (+G, parameter = 1.0457) to model the evolutionary diversities among sites. The tree was obtained using Maximum likelihood and the topology with the superior log likelihood value was selected. The evolutionary analysis was performed in MEGA7.Citation14
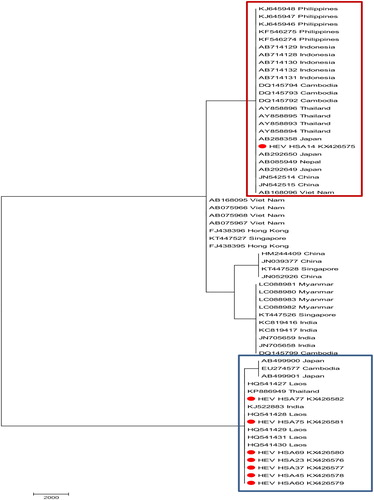
Interestingly, the Laos and Thailand isolates were from pigs and belong to genotypes 4 and 3, respectively, while the Indian isolate was from human serum and belongs to genotype 1 ().
Table 1 Genetic distribution of some Southeast Asian HEV isolates used in phylogenetic geographical relatedness with the Malaysian isolate. The Myanmar isolate genotype was not confirmed (–).
Malaysian isolates HSA14, 23 and 45 demonstrated high sequence identity with a swine isolate, SAAS-FX17 (JF915746), from China (96.7%, 99% and 94.1%, respectively), while isolate HSA75 was found to be 89.3% identical to swine isolate IND-SW-00-01 (AY723745) from India (). The evolutionary relatedness of these isolates as inferred by Neighbor-Joining demonstrated close genetic relatedness with swine and boars in a phylogenetic tree as shown in .
Fig. 3 Evolutionary relatedness of our Malaysian isolates (red dot) with swine and boar sequences inferred by Neighbor-Joining. The tree branches shown are optimal with a branch length sum of 4.43261981. The evolutionary diversities were computed via the Kimura 2-parameter based on the number of substitutions per site. The variation among sites was modelled by gamma distribution.
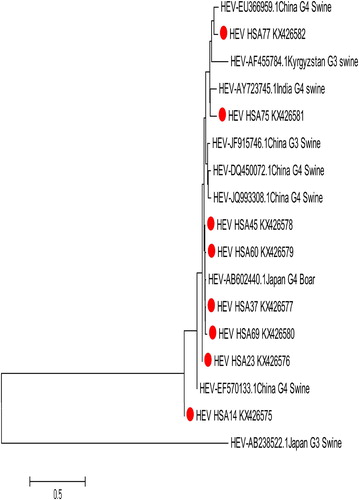
Table 2 Estimation of sequence evolutional divergence between our Malaysian isolates from humans and the reference isolates from animals. The Kimura 2-parameter model was used to analyse the number of base substitutions. The variation rate amongst the sequences was modelled with a gamma distribution and all gabs and missing data were eliminated. The evolutional diversity was conducted in MEGA7.Citation14 HEVHepatitis E virus, G Genotype.
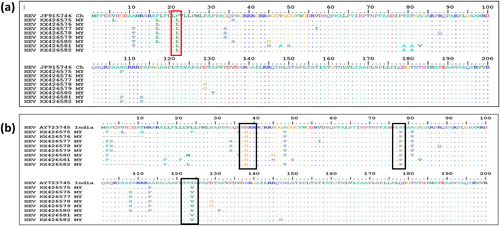
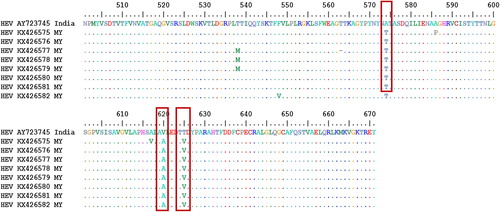
This evolutionary relationship was computed using the Kimura 2-parameter model with gamma distribution. Further bioinformatics analysis revealed that nucleotide substitution as well as transversion rates showed that transition and transversion occurred most with cytosine and thymine (25.11%), followed by substitutions between thymine and guanine (19.34%). Amino acid sequence analysis of our isolates compared to a reference swine HEV from China which exhibited 99% similarity with our sequences showed that our isolates had 3 to 11 amino acid variations from the swine HEV at different positions of the gene. However, all the Malaysian isolates shared an amino acid difference at position 22 of the ORF-2 gene where phenylalanine (F) in swine HEV was replaced by leucine (L) in human HEV (a). On the other hand, when compared with the Indian isolate with the lowest sequence similarity (89.3%), our Malaysian isolates showed more variations in amino acids than the China swine isolates but shared an amino acid variation at six different positions: D37G, S78P, T124V, A574T, V620A and T625V. The lowest number of amino acid differences from this Indian isolate was found in the Malaysian isolate KX426582, which differed from the Indian swine isolate by only 11 amino acids (b).
Fig. 4 Deduced amino acid alignments of the Malaysian (MY) human HEV isolate with a reference swine HEV: (a) the dots indicate common amino acids with the reference isolate. Reference swine isolate from China. The red box indicates a site of an amino acid difference shared by all of the Malaysian isolates (F22L) with the reference isolate. (b) The dots indicate common amino acids with the reference isolate. The black boxes indicate a site of an amino acid difference shared by all of the Malaysian isolates with the reference isolate. Where F = Phenylalanine, L = Leucine, T = Threonine, S = Serine, G = Glycin, A = Alanine, V = Valine, Q = Glutamine, N = Asparagine, P = Proline, I = Isoleucine, M = Methionine, Y = Tyrosine, D = Aspartic acid.
4 Discussion
This study provides strong evidence based on bioinformatics that HEV isolated from chronic hepatitis B patients in Malaysia has a strong link to swine hepatitis E. This suggests zoonotic transmission of HEV for the first time in Malaysia. Before now, only seroprevalence of HEV was reported from among Malaysian Orang Asli from Parit Tanjung and Betau which had a prevalence of 44 and 50%, respectively, while on the other hand the prevalence was lower among blood donors in Kuala Lumpur (2%).Citation19 However, in this study, HEV prevalence was found to be 9.8% among Malaysian chronic hepatitis B patients. All the eight HEV antibody positive samples were found to be positive for HEV RNA. This is a clear demonstration that the rate of infection within a single country can vary significantly, from 2% in urban blood donors to 50% in aboriginal populations and 9.8% in chronic hepatitis B patients as found in this study. This suggests the need for a comprehensive study to evaluate the prevalence of HEV in general population for effective control measures. Similarly, in Netherland, an 11% HEV prevalence was found in those exposed to swine, with a prevalence of only 2% in those not exposed,Citation20 indicating the significance of exposure to swine. Several studies reported a higher HEV prevalence in males compared to females, such as 49.8% and 44.9% in Nepal and 52.6% and 47.4% in Bangladesh.Citation21 Similarly, our results also revealed a higher prevalence in males (62.5%) than in females (37.5%). Most of the patients with HEV in this study were 61 years of age and older (75%), which is in agreement with the findings from previous studies.Citation12,Citation22 The racial aligning of HEV to Chinese in this study might not be surprising due the fact that they consume pork. Pigs, wild boar and rats have been shown experimentally to be associated with HEV strain that affects humans.Citation23–Citation25 Swine hepatitis E virus has since been discovered to cross reacts with human HEV antibodies.Citation26 This significantly raises the public health concern for a zoonotic transmission, which is greatly supported by the finding in this study which shows that the Malaysian human HEV isolates have high sequence identity (89–99%) to swine and boar HEV. Similarly, the zoonotic potential of HEV has also been reported in the United States of America,Citation27 which shows that non endemic countries can also have zoonotic transmission of HEV.Citation28
In this study, the phylogenetic analysis was based on the ORF-2 nucleotides and confirmed for the first time that the Malaysian isolates were of genotype 4. Studies have shown that genotype 3 and 4 have zoonotic reservoir while genotype 1 and 2 are exclusively human pathogens.Citation29 Similarly, genotypes 1 and 4 were found in China,Citation30 genotype 2 in MexicoCitation31 and NigeriaCitation32 and genotype 3 the United States of America.Citation33 However, Malaysian isolates HSA14, 23 and 45 revealed high sequence identity with a swine isolate, SAAS-FX17 (JF915746), from China (96.7%, 99% and 94.1%, respectively) which also belongs to genotype 4. Similarly, isolate HSA75 was also found to be 89.3% identical to swine isolate (AY723745) of India and also belong to genotype 4. When the amino acid analysis of our Malaysian isolate was compared with the China swine isolate that had the highest identity (99%) to our isolate, it was found that they only differed by an average of eight amino acids. This shows how close the Malaysian HEV isolate is with the China swine isolate JF915746. The amino acid variation that is common to all of our Malaysian isolates was found to be at position 22, where leucine was present in the Malaysian human isolate while phenylalanine was present in the China swine isolates. On the other hand, an Indian swine isolate AY23745 with the least (89.3%) similarity to our Malaysian human isolate showed more amino acid variation at various positions of the ORF-2 protein. The lowest number of amino acid differences was found in Malaysian isolate KX426582, which differed from the Indian swine HEV by just eleven amino acids. All the Malaysian isolates shared amino acid differences with the Indian swine isolates at the following positions: at 37 swine aspartic acid was replaced by glycine in human HEV (D37G), at 78 swine serine was replaced by proline in human HEV (S78P), at 124 swine threonine was replaced by valine in human HEV (T124V), at 574 swine alanine was replaced by threonine in human HEV (A574T), at 620 swine valine was replaced by alanine in human HEV (V620A) and at 625 swine threonine was replaced by valine in human HEV, as shown in .
Fig. 5 Deduced amino acid alignments of the Malaysian isolate with a reference swine HEV capsid protein from India. The red boxes indicate a site of shared amino acid of the Malaysian isolates with the reference swine isolate. Where F = Phenylalanine, L = Leucine, T = Threonine, S = Serine, G = Glycine, A = Alanine, V = Valine, Q = Glutamine, N = Asparagine, P = Proline, I = Isoleucine, M = Methionine = Tyrosine.
Although these Malaysian isolates resemble the Chinese and Indian isolates, all were from Malaysians whose history of travel to China and India was not ascertained. Nonetheless, it is still possible that these isolates were imported from China and India, because of importation of many possible vehicles of transmission such as canned foods, pork meats as well as immigration of people from these and other Asian countries to Malaysia for tourism or business. A wider understanding of the risk and prevalence of HEV associated with swine in the Southeast Asia region could be of public health importance and essential in food safety intervention.
5 Conclusion
This study revealed that 9.8% of chronically infected HBV patients were co-infected with HEV. Comparative genomic sequence analysis of our isolates suggests a zoonotic origin, with sequence identity ranging from 89% to 99% with swine and boar HEV. Since HEV is capable of causing fulminant hepatic failure in patients with pre-existing liver diseases, it will therefore be of significant benefit to screen all chronic hepatitis B patients of HEV co-infection in order to provide effective patient management and disease prevention.
Sources of support
This work was supported by the Fundamental Research Grant Scheme (FRGS) from Ministry of Higher Education, Malaysia, Grant No. 5524778.
Conflict of interest
None to declare.
Notes
Peer review under responsibility of Alexandria University Faculty of Medicine.
Available online 9 October 2017
References
- H.R.DaltonW.K.SaundersMHepatitis E virus in developed countries: one of the most successful zoonotic viral diseases in human historyJ Virus Erad120152329
- N.KamarH.R.DaltonF.AbravanelJ.IzopetHepatitis E virus infectionClin Microbiol Rev272014116138
- E.Delarocque-AstagneauF.AbravanelA.MoshenEpidemiological and virological characteristics of symptomatic acute hepatitis E in Greater Cairo, EgyptClin Microbiol Infect182012982988
- J.AmbrosioniA.MaminA.HadengueLong-term hepatitis E viral load kinetics in an immunocompromised patient treated with ribavirinClin Microbiol Infect2020140718720
- R.AggarwalS.JameelHepatitis eHepatology54201122182226
- K.FujiwaraS.YasuiY.YonemitsuImportance of the poor prognosis of severe and fulminant hepatitis in the elderly in an era of a highly aging society: analysis in a Japanese centerHepatol Res452015863871
- X.-J.MengZoonotic and xenozoonotic risks of the hepatitis E virusInfect Dis Rev220003541
- S.N.ChatterjeeP.B.DevhareS.Y.PingleM.S.PaingankarV.A.ArankalleK.S.LoleHepatitis E virus (HEV)-1 harbouring HEV-4 non-structural protein (ORF1) replicates in transfected porcine kidney cellsJ Gen Virol97201618291840
- S.TeiN.KitajimaK.TakahashiS.MishiroZoonotic transmission of hepatitis E virus from deer to human beingsLancet3622003371373
- Y.YazakiH.MizuoM.TakahashiSporadic acute or fulminant hepatitis E in Hokkaido, Japan, may be food-borne, as suggested by the presence of hepatitis E virus in pig liver as foodJ Gen Virol84200323512357
- H.MatsudaK.OkadaK.TakahashiS.MishiroSevere hepatitis E virus infection after ingestion of uncooked liver from a wild boarJ Infect Dis1882003944
- A.R.GarbugliaA.I.AlessandriniN.PavioMale patient with acute hepatitis E in Genoa, Italy: figatelli (pork liver sausage) as probable source of the infectionClin Microbiol Infect212015e4e6
- F.RenC.ZhaoL.Wangvirus seroprevalence and molecular study among blood donors in ChinaTransfusion542014910917
- S.KumarG.StecherK.TamuraMEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasetsMol Biol Evol2016
- M.NeiS.KumarMolecular evolution and phylogenetics2000Oxford University Press
- J.FelsensteinConfidence limits on phylogenies: an approach using the bootstrapEvolution391985783791
- Hall TA, editor. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic acids symposium series1999.
- M.HasegawaH.KishinoT.-A.YanoDating of the human-ape splitting by a molecular clock of mitochondrial DNAJ Mol Evol221985160174
- H.-F.SeowN.MahomedJ.-W.MakM.A.RiddellF.LiD.A.AndersonSeroprevalence of antibodies to hepatitis E virus in the normal blood donor population and two aboriginal communities in MalaysiaJ Med Virol591999164168
- M.BouwknegtB.EngelM.HerremansBayesian estimation of hepatitis E virus seroprevalence for populations with different exposure levels to swine in The NetherlandsEpidemiol Infect1362008567576
- J.IzopetA.B.LabriqueB.BasnyatHepatitis E virus seroprevalence in three hyperendemic areas: Nepal, Bangladesh and southwest FranceJ Clin Virol7020153942
- WK.PaverPP.MortimerAn overview of the hepatitis virusesClin Microbiol Infect21996132141
- M.BalayanR.UsmanovN.ZamyatinaD.DjumalievaF.KarasExperimental hepatitis E infection in domestic pigsJ Med Virol3219905859
- Y.ManeeratE.T.ClaysonK.S.MyintG.D.YoungB.L.InnisExperimental infection of the laboratory rat with the hepatitis E virusJ Med Virol481996121128
- X.-J.MengP.G.HalburM.S.ShapiroGenetic and experimental evidence for cross-species infection by swine hepatitis E virusJ Virol72199897149721
- X.-J.MengR.H.PurcellP.G.HalburA novel virus in swine is closely related to the human hepatitis E virusProc Natl Acad Sci94199798609865
- G.G.SchlauderG.J.DawsonJ.C.ErkerThe sequence and phylogenetic analysis of a novel hepatitis E virus isolated from a patient with acute hepatitis reported in the United StatesJ Gen Virol791998447456
- L.ChristouM.KosmidouHepatitis E virus in the Western world–a pork-related zoonosisClin Microbiol Infect192013600604
- X.MengHepatitis E virus: animal reservoirs and zoonotic riskVet Microbiol1402010256265
- Y.WangR.LingJ.C.ErkerA divergent genotype of hepatitis E virus in Chinese patients with acute hepatitisJ Gen Virol801999169177
- C.-C.HuangD.NguyenJ.FernandezMolecular cloning and sequencing of the Mexico isolate of hepatitis E virus (HEV)Virology1911992550558
- Y.BuissonM.GrandadamE.NicandIdentification of a novel hepatitis E virus in NigeriaJ Gen Virol812000903909
- D.L.SmalleyM.F.HallC.L.BroughtonAutoantibodies present in chronic hepatitis C and chronic hepatitis B viral infectionsHepatology271998 1452-1452
