Abstract
Early diagnosis of infectious diseases represents powerful means to increase patient survival rate, avoid disease spreading, and decrease healthcare costs. Current Polymerase Chain Reaction (PCR)- and antibody-based diagnostic methods for detecting pathogens offer rapid analysis with highly accurate and specific results. However, those methods are still hampered by the need of sophisticated infrastructures and highly-skilled technicians, which limit the deployment in developing area. Synthetic biology with its rational and short design-to-production cycles has the potential to overcome those limitations. Here, we discuss two promising efforts for pathogen nucleic acids detection using synthetic biology approaches: Synthetic RNA-based and Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated (CRISPR/Cas)-based biosensors. The two systems were reported to show remarkable specificity and sensitivity on detecting and reporting the presence of pathogen via pathogen nucleic acid recognition with lower development and operational costs when compared to current PCR- and antibody-based diagnostic tools. Moreover, both systems can be applied to paper-based platforms which simplify the distribution and utilization in low resource-settings.
1 Introduction
In vitro diagnostic (IVD) test, in which examination is performed on specimens taken from the human body, is an indispensable tool for producing high-quality medical outcomes.Citation1,Citation2 Such tests are critical for diagnosing and monitoring of diseases, providing prognosis and predicting treatment response, as well as, assessing the potential risk of developing a disease and guiding patient management.Citation2
Currently there are more than 40,000 types of IVD test available with global economic values forecasted to reach $75 billion US Dollar (USD) by 2020.Citation3 In general, those wide varieties of IVD test can be categorized in three main types: (1) Clinical laboratory tests: Relatively complex tests, where certain samples from patients are sent to clinical laboratories for examination using advanced laboratory facilities by skilled operators (i.e. PCR-based and immunoassay / antibody-based testing for pathogen detection). (2) Near-patient tests: Relatively simple tests which does not rely on sophisticated instruments and can be easily performed on a simple platform by physicians or nurses (i.e. urine test strips for determining pH, specific gravity, and the presence of certain chemicals). (3) In-home tests: The simplest tests, where patients can do the tests and deal with the test results information by themselves (i.e. diabetic glucose meters and home pregnancy test).Citation1
Particularly in developing countries, IVD test plays important role on global efforts to combat infectious diseases.Citation4 It was reported in 2007; early in vitro diagnosis could have eliminated as many as 92,000 deaths from bacterial infection and reduced healthcare costs by at least $1.5 billion in the United States.Citation5 While in the other reports, early diagnosis and treatment of Tuberculosis (TB) have saved approximately 43 million lives from 2000 to 2014.Citation6 In general, IVD test represents a powerful way to optimize treatment, increase patient survival rate, and decrease healthcare costs.
Among the most commonly used IVD tests for infectious diseases are culturing, biomarker identification by PCR and immunoassays, and whole genome sequencing.Citation4 Despite the specificity and sensitivity, the application of those tests is still hampered by various problems, mainly related to analysis time, cost, and portability for a simple in-field application. Culturing represents the simplest and economical diagnosis methods, however in some cases such as TB, the growth of the pathogens is remarkably slow which might prevents patients for receiving timely and adequate treatments.Citation7 On the other hand, biomarker identification and whole genome sequencing offers fast and accurate analysis; however, those methods are costly and requires highly trained experts, which makes deployment in area with poor infrastructures challenging. Thus, novel IVD approaches need to be developed to meet those demands.
Recently, the emerging field of synthetic biology is employed to tackle global challenges on developing low-cost, rapid, sensitive, easy to use, and adaptable high clinical value diagnostic tools.Citation8–Citation10 In this review we will highlight two synthetic biology-based IVD systems, synthetic RNA- CRISPR/Cas-based biosensors which have been demonstrated to detect and report various nucleic acid biomarkers from pathogens with high accuracy and sensitivity, relatively simple logistics, and low development and operational costs. The future direction of using both systems as infectious disease diagnostic platforms will also be discussed.
2 Synthetic biology-based in vitro diagnostic approaches
Synthetic biology utilizes forward engineering approaches to seek interchangeable parts from natural biology and assemble those parts into systems that function unnaturally.Citation9 In the context of development of diagnostic methods, synthetic biology approaches are typically focused on building novel biosensing systems with a modular architecture consisting of sensor, signal processor, and reporter modules with measurable output ().Citation11 As synthetic biology field matured, most components and parts to build such biosensing system are readily standardized and catalogued.Citation12 Consequently, when applied in the development of IVD platforms, the utilization of synthetic biology approaches is relatively more straightforward and inexpensive when compared to PCR-based and antibody-based platforms.Citation4,Citation9
Fig. 1 Synthetic biology-based in vitro diagnostic system. Synthetic biology enables standardization and re-engineering of biological parts involved in sensing and processing biological signals to form synthetic diagnostic system, consisting of sensor, signal processor, and reporter modules, which meet clinical specifications to aid medical decision-making.Citation11
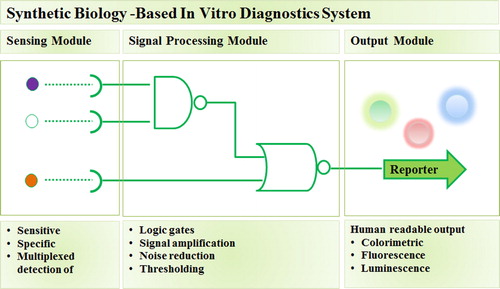
Here, we focus on two promising synthetic biology-inspired IVD platforms, Synthetic RNA- and CRISPR/Cas-based biosensors. Both methods detect and report the presence of pathogen via pathogen nucleic acid recognition. Synthetic RNA biosensor sensing module consists of RNA switch which contain sequences complementary to the target pathogen RNA. The binding of target RNA will activate the expression of reporter gene which product can be observed visually. While the CRISPR/Cas-based systems involve CRISPR RNA that bind to pathogen DNA or RNA and activate the non-specific activity of Cas nuclease to cleave quenched fluorescent reporter RNA. The cleaved RNA reporter will emit fluorescence signal which can be detected easily. Furthermore, the two systems can be applied to a paper-based platform which reduce operational costs, as well as simplify the storage and deployment in area with lack of advanced infrastructures and medical experts.
3 Synthetic RNA-based biosensors
RNA molecules are able to take on complex structures defined by their sequence and can mediate diverse modular functions across distinct sequence domains.Citation13,Citation14 Moreover, RNA molecules can act as genetic regulatory elements at the level of transcription or translation.Citation14,Citation15 Those aforementioned features of RNAs are well-suited for rational design, and ultimately can be engineered as biosensor, which offers wide dynamic range, low system crosstalk, and design flexibility.Citation16
RNA-based biosensors consist of genetic circuits with riboregulators that act as a switch (A).Citation17 The genetic circuit would be activated, expressing the reporter gene, upon the binding of target biomarker RNA to the riboregulator. Until recently, riboregulators relied on cis-repression sequence to sequester the ribosomal binding site (RBS) for controlling the translation of the downstream mRNA. In the presence of higher affinity RNA molecule (the transactivator), the RBS is exposed, and translation ensues. The application of RNA-based biosensors with this conventional riboregulator is very limited, mainly due to the sequence constraints of the transactivator (target biomarker RNA), which need to contain the RBS sequence.Citation9
Fig. 2 RNA-based biosensors. (A) Conventional riboregulators control the translation of downstream RNA by sequestering the RBS through a cis-repression sequence, which is relieved in the presence of the transactivator RNA (taRNA). (B) Toehold switch, in which RBS is located in a hairpin loop within the repressed RNA’s 5′ untranslated region and released upon the binding of trigger RNA to the toehold, unzipping the hairpin into linear structures, releasing the RBS and start codon (AUG).Citation17
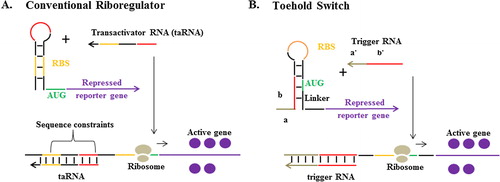
The alternative design for the riboregulator was reported by Green et al.Citation17 in the form of toehold switches (B). In their design the need for sequence conservation between the transactivator and the RBS is eliminated by engineering “toehold” sequence for the binding of the target biomarker RNA (trigger RNA). Translation of the reporter gene begins only when trigger RNA binds to the toehold, which unzips the hairpin into linear structures, releasing the RBS and start codon. Moreover, this novel riboregulator can be engineered to detect not only short trigger RNA, but also full length mRNA, thus offering wider range of application when used as a component of RNA-based biosensors.Citation18
Smart design of in vitro diagnostic device based on toehold switches mRNA sensors, was reported by Pardee et al.Citation18 In this arrangement, appropriate amount of plasmid encoding the synthetic gene networks and commercially available or in-house cell-free transcription system (RNA polymerase, energy source (Adenosine triphosphate (ATP), transcription factors, and nucleotides) and translation system (Ribosome, energy sources (ATP and Guanidine triphosphate (GTP), tRNA, and Amino acids) are freeze dried onto paper or other porous materials ().Citation18–Citation20 Upon rehydration the RNA-based biosensor will be transcribed using the plasmid as template, and in the presence of target RNA the reporter gene will be translated. This IVD device is stable for long-term storage at room temperature.
Fig. 3 Portable synthetic RNA biosensor-based diagnostic devices. Diagnostic genetic circuit controlled by toehold switch and cell free coupled transcription/translation system are assembled into paper or other porous materials. The genetic circuit is activated when rehydrated and exposed to the test sample containing target RNAs or small molecules.Citation18
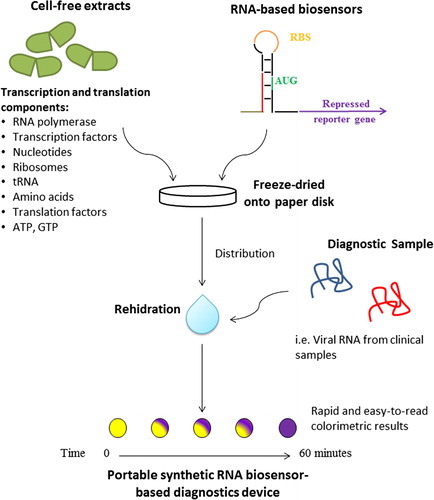
This paper-based system demonstrated promising diagnostic application by detecting mRNA of various antibiotic resistance genes. Moreover, the system successfully distinguished two Ebola virus strains, Zaire and Sudan, which differs in length by only three nucleotides, with the limit of Ebola nucleoprotein mRNA detection of 30 nanomolar (nM), demonstrating its high specificity and sensitivity.Citation18,Citation20
The paper-based Synthetic Gene Networks platform showed promising advantages as a diagnostic tool, as it can be easily and economically mass produced, stored, operated, and deployed, especially in the area where laboratory infrastructures is not available.Citation19 This paper-based system can cost as little as $0.02 to $0.04 USD per sensor using in-house cell-free expression system (or $0.35 to $0.65 USD using commercial cell-free expression systems), compared to $0.45 to $1.40 USD for a single antibody-based rapid detection test reaction and $1.50 to $4.00 USD (reagents only) for PCR-based test.Citation18,Citation20 In term of development, this system represents significantly lower development costs and shorter design to production cycles when compared to antibody-based diagnostics. Toehold switch mRNA sensors using in the paper-based system can be constructed in less than 12 h with the cost of $21 USD/sensor, which are more favorable than 2–6 months development time and $4000-$30,000 USD development cost for typical custom commercial antibody production.Citation18,Citation20 Moreover, the Toehold switch mRNA sensors can be designed based on the genetic information of pathogen alone, which is ideal for the development of diagnostic tools for emerging pathogens.
4 CRISPR/Cas-based biosensors
4.1 First-generation CRISPR/Cas-based biosensors
Microbial Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated (CRISPR/Cas) adaptive immune systems contain programmable endonucleases that can recognize specific nucleic acid sequences.Citation21 One class of Cas enzyme, Cas13a, can be reprogrammed to target specific RNA sequence when work in concert with CRISPR RNAs (crRNAs)Citation22,Citation23 (A). Upon recognition of its RNA target as specified by the crRNA sequence, activated Cas13a engages in “collateral” cleavage of nearby non-targeted RNAs, regardless of homology to the crRNA.Citation24 Those characteristics of CRISPR-Cas13 system can be harnessed to construct CRISPR-based diagnostics platform (C).
Fig. 4 Cas endonuclease activity and nucleic acid detection systems. Cas endonucleases are single protein effector for CRISPR RNA which bind and cleave the target (A) RNA, in the case of Cas13a, and (B) DNA, in the case of Cas12a, complementary to the CRISPR RNA. Upon target detection collateral cleavage of single-stranded RNA (ssRNA; Cas13) or single-stranded DNA (ssDNA; Cas12a) is activated. (C) The collateral cleavage activity can be utilized to cleave and activate a quenched fluorescent reporter RNA, which indicating the presence of the target nucleic acids of interest.
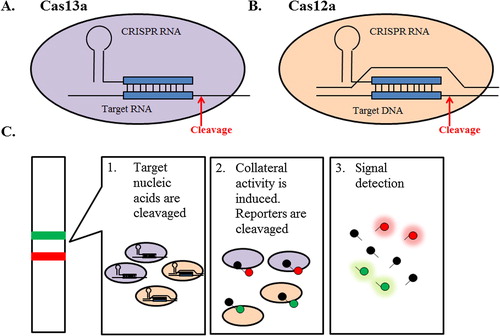
Such platform was developed and termed as SHERLOCK (Specific High-Sensitivity Enzymatic Reporter unLOCKing).Citation24 In SHERLOCK, the collateral cleavage activity is harnessed to cleave and activate a quenched fluorescent reporter RNA, which indicating the presence of the target RNA of interest. SHERLOCK is able to detect target RNA on attomolar (10−18 mol/L) scale which is more sensitive than currently reported PCR-based method with the detection limit on femtomolar (10−15 mol/L) scale.Citation25 Such remarkable sensitivity is achieved using highly active Cas13a ortholog from Leptotrichia wadei (LwCas13a); and the incorporation of a recombinase polymerase amplification (RPA) step coupled with T7-mediated transcription for target enrichment prior to detection.
The sensitivity and specificity of SHERLOCK was demonstrated by its ability to detect target viral RNA down to concentrations of ∼2 attomolar (aM) and discriminate between Zika virus (ZIKV) and the related Dengue virus (DENV).Citation24,Citation26 Furthermore, by specifically design the crRNAs, SHERLOCK showed remarkable feat on distinguishing different strains of the same virus, including the African and American strains of ZIKV, and strain 1 and 3 of DENV, in which genetic material only differs by few bases. In additional demonstration, SHERLOCK was also able to correctly detect different genotypes of pathogenic bacterial strains such as E. coli and Pseudomonas aeruginosa, with low cross-reactivity and detect antibiotic resistance genes from various clinical isolates of Klebsiella pneumoniae.Citation24
4.2 Second-generation CRISPR/Cas-based biosensors
To further expands the functionality and application of CRISPR/Cas-based biosensors, extensive researches on Cas endonuclease mechanisms was conducted. Recently, it was discovered that Cas12a proteins cleave double-stranded DNA (dsDNA) in a sequence-specific manner (B). The proteins induce robust collateral cleavage against nonspecific single-stranded DNA (ssDNA).Citation27 A diagnostic platform then was developed by coupling Cas12a-based DNA reporter with isothermal pre-amplification by recombinase polymerase amplification (RPA), termed as DETECTR (DNA Endonuclease Targeted CRISPR Trans Reporter). DETECTR was successfully applied for rapid and accurate qualitative detection carcinoma-associated Human Papilloma Virus (HPV) types 16 and 18 from clinical specimens, and showed 1 attomolar detection limit when tested against synthetic plasmids containing cognate target sequences.Citation27
Meanwhile, SHERLOCK also underwent substantial upgrade in term of sensitivity and functionality by introducing various new approaches in the system ().Citation28 In sample preparation step subsequent heating at 37–50 °C for 5–20 min and 64–95 °C for 5 min termed as HUDSON (Heating Unextracted Diagnostic Samples to Obliterate Nucleases), was employed.Citation28,Citation29 This method ensures the release of target viral nucleic acids from clinical specimens, bypassing the need for nucleic acid extraction, and protects them from degradation. SHERLOCK coupled with HUDSON permitted sensitive detection of ZIKV RNA from infectious particles at up to 0.9 aM in clinical sample in less than 2 h, which is comparable to ZIKV RNA concentrations observed in patient samples, which range from 0.9 to 900 aM.Citation29
Fig. 5 CRISPR/Cas-based diagnostics workflow. Target nucleic acids was obtained by HUDSON, then amplified by isothermal amplification to achieve attomolar sensitivity. The resulting DNA can be transcribed for detection using SHERLOCK or detected directly by DETECTR and SHERLOCKv2.Citation28
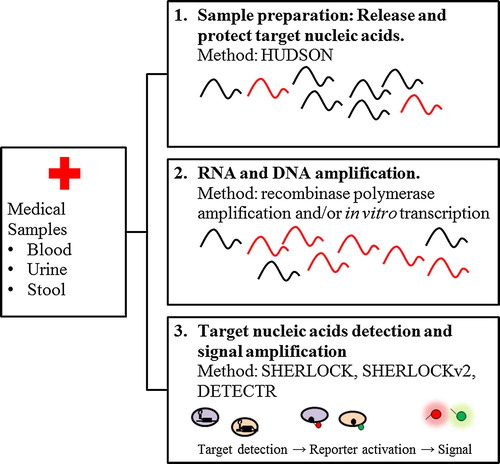
The extensive characterization of 17 CRISPR-Cas13a and -Cas13b orthologs, along with the discovery of collateral cleavage activity of Cas12a, enabled the development of SHERLOCK version 2 (v2). Three CRISPR13 orthologs and Cas12a was combined and coupled RPA to form a platform that allows for detection of three ssRNA targets and one dsDNA target in a single reaction. It was demonstrated that SHERLOCKv2 accurately detected ZIKV ssRNA, synthetic ssRNA, DENV ssRNA, and synthetic dsDNA by visual readout in less than 90 minutes.Citation30
To further boost the signal output, nonspecific RNAse Csm6 was incorporated into the SHERLOCK v2 platform with dual reporter system which consist of poly-adenylate (polyA) and quenched fluorescent RNA reporters were used in the platform. Upon recognition of the target RNA, Cas13a collateral activities cleaves PolyA reporter, the cleaved product then will activate Csm6. Cas13a and Csm6 then work in concert to cleave quenched fluorescent RNA reporter, increase the signal of SHERLOCKv2 by up to 3.5-folds.Citation30
Finally, for in-field clinical applications, the reaction reagents of CRISPR/Cas-based biosensors; Cas protein-CRISPR RNA complex and RNA reporters can be lyophilized in a paper-based platform for long-term storage, and easy deployment. Specific and sensitive paper-based CRISPR/Cas-based biosensors can be designed and synthesized in a matter of days for as low as $0.61 USD/test.Citation24
5 Conclusion
As demonstrated by portable synthetic RNA-based and CRISPR/Cas-based biosensors, synthetic biology approaches on developing in vitro diagnostics platform enable short design to production cycles, and simultaneous advantages of low-cost, rapid, and highly accurate analysis. Furthermore, both systems offer logistical advantages compared to commonly use PCR-based nucleic acid detection tests when deployed to developing areas, where infrastructures and skilled personnel are limited, and more importantly, where delays in diagnosis, targeted care, and infection control might contribute to infectious disease mortality and spread.Citation31
Among the two systems, portable synthetic RNA-based biosensor offers lower cost and simpler application since enrichment steps as in CRISPR/Cas-based biosensors are not needed. Meanwhile, CRISPR/Cas-based biosensors system offers higher sensitivity with higher operational cost and more complex in-field application. Continued researches on both synthetic biosensors would undoubtedly uncover more useful RNA switches and enzymes which might be implemented for improved sensitivity of both systems. In the case of CRISPR/Cas-based biosensors, those may eliminate the needs for enrichment step, which would simplify the testing procedures and reduce the analysis cost. Nevertheless, in both systems, clinical testing and trials, including benchmarking against existing diagnostic tools, are required to ensure the quality of the analysis results.Citation26 Additionally, the effectiveness of RNA-based and CRISPR/Cas-based biosensors needs to be evaluated in in-field clinical situation, where environmental pressures such as high ambient temperatures or dust might reduce the test performances.Citation32
Finally, multi-target detection features on both systems needs to be further developed and optimized, considering the importance of co-infection diagnosis. One important example is pneumonia, the leading killer of children worldwide, which caused by DNA and RNA viruses alone or in combination with bacterial infection.Citation33 Another example is secondary infection on HIV patient which might involve bacteria (TB, pneumonia), DNA virus (Hepatitis B virus), or RNA virus (Hepatitis C virus).Citation34
Conflict of interest
The authors declared that there is no conflict of interest.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Notes
Peer review under responsibility of Alexandria University Faculty of Medicine.
Available online 13 December 2018
References
- W.ZhouX.GaoD.LiuX.ChenGold nanoparticles for in vitro diagnosticsChem Rev1151920151057510636
- U.P.RohrC.BinderT.DieterleThe value of in vitro diagnostic testing in medical practice: a status reportPLoS ONE1132016e0149856
- Y.LokkoM.HeijdeK.SchebestaP.ScholtesM.Van MontaguM.GiaccaBiotechnology and the bioeconomy – towards inclusive and sustainable industrial developmentN Biotechnol40Pt A2018510
- T.Y.WeiC.M.ChengSynthetic biology-based point-of-care diagnostics for infectious diseaseCell Chem Biol239201610561066
- A.F.ShorrS.T.MicekW.L.JacksonJr.M.H.KollefEconomic implications of an evidence-based sepsis protocol: can we improve outcomes and lower costs?Crit Care Med355200712571262
- B.KhwannimitR.BhurayanontachaiThe direct costs of intensive care management and risk factors for financial burden of patients with severe sepsis and septic shockJ Crit Care3052015929934
- C.M.DenkingerS.V.KikD.M.CirilloDefining the needs for next generation assays for tuberculosisJ Infect Dis211Suppl 22015S29S38
- D.E.CameronC.J.BashorJ.J.CollinsA brief history of synthetic biologyNat Rev Microbiol122014381
- S.SlomovicK.PardeeJ.J.CollinsSynthetic biology devices for in vitro and in vivo diagnosticsProc Natl Acad Sci USA1124720151442914435
- D.BraffD.ShisJ.J.CollinsSynthetic biology platform technologies for antimicrobial applicationsAdv Drug Deliv Rev10520163543
- A.CourbetE.RenardF.MolinaBringing next-generation diagnostics to the clinic through synthetic biologyEMBO Mol Med892016987991
- B.CantonA.LabnoD.EndyRefinement and standardization of synthetic biological parts and devicesNat Biotechnol2672008787
- F.J.IsaacsD.J.DwyerC.DingD.D.PervouchineC.R.CantorJ.J.CollinsEngineered riboregulators enable post-transcriptional control of gene expressionNat Biotechnol222004841
- C.MyhrvoldP.A.SilverUsing synthetic RNAs as scaffolds and regulatorsNature Struct Amp Mol Biol2220158
- J.ChappellK.E.WattersM.K.TakahashiJ.B.LucksA renaissance in RNA synthetic biology: new mechanisms, applications and tools for the futureCurr Opin Chem Biol2820154756
- J.B.LucksL.QiV.K.MutalikD.WangA.P.ArkinVersatile RNA-sensing transcriptional regulators for engineering genetic networksProc Natl Acad Sci USA10821201186178622
- A.A.GreenP.A.SilverJ.J.CollinsP.YinToehold switches: De-Novo-designed regulators of gene expressionCell15942014925939
- K.PardeeA.A.GreenT.FerrantePaper-based synthetic gene networksCell15942014940954
- A.J.LopatkinL.YouSynthetic biology looks good on paperCell15942014718720
- K.PardeeA.A.GreenM.K.TakahashiRapid, low-cost detection of Zika virus using programmable biomolecular componentsCell1655201612551266
- R.BarrangouC.FremauxH.DeveauCRISPR provides acquired resistance against viruses in prokaryotesScience3155819200717091712
- G.J.KnottA.East-SeletskyJ.C.CofskyGuide-bound structures of an RNA-targeting A-cleaving CRISPR–Cas13a enzymeNature Struct Amp Mol Biol242017825
- O.O.AbudayyehJ.S.GootenbergP.EssletzbichlerRNA targeting with CRISPR–Cas13Nature55076752017280
- J.S.GootenbergO.O.AbudayyehJ.W.LeeNucleic acid detection with CRISPR-Cas13a/C2c2Science35663362017438442
- J.DongG.ChenW.WangColorimetric PCR-based microRNA detection method based on small organic dye and single enzymeAnal Chem2018
- D.G.SashitalPathogen detection in the CRISPR-Cas eraGenome Med101201832
- J.S.ChenE.MaL.B.HarringtonCRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activityScience36063872018436439
- D.S.ChertowNext-generation diagnostics with CRISPRScience36063872018381382
- C.MyhrvoldC.A.FreijeJ.S.GootenbergField-deployable viral diagnostics using CRISPR-Cas13Science36063872018444448
- J.S.GootenbergO.O.AbudayyehM.J.KellnerJ.JoungJ.J.CollinsF.ZhangMultiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6Science36063872018439444
- R.W.PeelingP.G.SmithP.M.M.BossuytA guide for diagnostic evaluationsNat Rev Microbiol82010S2
- A.NiemzT.M.FergusonD.S.BoylePoint-of-care nucleic acid testing for infectious diseasesTrends Biotechnol2952011240250
- J.A.McCullersThe co-pathogenesis of influenza viruses with bacteria in the lungNat Rev Microbiol122014252
- L.MilazzoS.AntinoriHepatitis virus and HIV interactionsLancet Infect Dis1411201410251027
