Abstract
There are three species of the genus Mesembryanthemum recorded in Egyptian flora (Mesembryanthemum crystallinum L., Mesembryanthemum nodiflorum L. and Mesembryanthemum forsskaolii Hochst. ex Bioss.). Various genetic markers (SDS-PAGE, RAPD and ISSR) have been applied to evaluate the genetic variability and structure within and between twelve populations of Egyptian Mesembryanthemum L. species collected from different localities along the International Coastal Road. The electrophoretic analysis of protein was revealed 18 bands of molecular weight ranging from 11.6 to 104.6 KDa. Five RAPD markers generated 39 total bands. The percentage of polymorphism ranged from 25% to 75%, with an average 61.78%. A total of five ISSR primers produced 37 total bands and the percentage of polymorphism ranged from 25% to 55.5%, with an average 40.77%. Results of AMOVA analysis showed that the percentages of molecular variance within populations are 73%, 77% and 53% and among species populations were scored as 27%, 23% and 47% using SDS-PAGE, RAPD and ISSR markers respectively. The results of the study revealed that M. crystallinum and M. nodiflorum exhibited within range genetic diversity. RAPD and ISSR markers exhibited lower genetic diversity for M. forsskaolii. Comparing with native species at local scale, M. crystallinum and M. nodiflorum had lower genetic diversity.
1 Introduction
Subfamily Mesembryanthemoideae, comprises the single genus Mesembryanthemum, with 102 species [Citation1]. The genus comprises about 70 species and it is native to the Mediterranean region, Atlantic Islands, Saudi Arabia, South Africa, South Australia and California [Citation2]. Three species of the genus Mesembryanthemum are recorded in Egyptian flora (Mesembryanthemum crystallinum L., Mesembryanthemum nodiflorum L. and Mesembryanthemum forsskaolii Hochst. ex Bioss.)
According to El-Shayeb et al. [Citation3], M. crystallinum grows in sandy and loamy habitats near the sea that are characterized by high soil moisture, moderate rainfall and temperature. M. forsskaolii grows in habitats of sandy plains or slopes that are characterized by low rainfall and high temperature. M. nodiflorum grows in wide ecological amplitude in sandy habitats that are characterized by low rainfall, high temperature and silty and loamy habitats with moderate rainfall and temperature. M. crystallinum and M. nodiflorum may grow in salty soil while, M. forsskaolii grows in the soil with low salinity.
Several studies have investigated Aizoaceae species such as M. crystallinum which has many biological activities (e.g. antioxidant, antiviral and antimicrobial) [Citation4Citation[5]–Citation6]. M. crystallinum may be also useful for bio-remediation due to its salt accumulation property [Citation7]. M. forsskaolii seeds were ground into flour used as a replacement for wheat for bread and cookies due to containing high protein content [Citation8]. Moreover, M. nodiflorum as well as M. crystallinum were used as food and in Tunisian traditional medicine for treatments of ocular infection [Citation9], making these three species good candidates for pharmaceutical or cosmetical applications.
The primary concern in population and evolutionary genetic studies is to assess and evaluate the genetic variation that it provides the basis for any evolutionary change. The methods for estimating genetic diversity have been progressed over the years from Mendelian analyses of morphological traits, to statistical characterization of quantitative characters, to variants biochemical electrophoretic assays and, recently, to molecular estimations of DNA sequence variation [Citation10].
The application of molecular genetics methods for assessing genetic diversity has been a significantly increased. They have been applied to increase our knowledge of the structure and extent of genetic variation within and among species [Citation11]. Several laboratory techniques had been developed including protein methods such as isozyme electrophoresis or molecular methods such as DNA analysis giving a high indication of the levels of genetic variation found in a species or population without direct interference from environmental factors [Citation11,Citation12].
Random Amplified Polymorphic DNA (RAPD) is one of the DNA techniques which can detect the polymorphism in closely related organism [Citation13]. Inter simple sequence repeat (ISSR)-PCR marker is also highly polymorphic and is useful in studies on genetic diversity, genome mapping, phylogeny, evolutionary biology, and population and conservation genetics [Citation14,Citation15]. Unlike allozymes which are codominant markers, RAPD and ISSR are dominant markers [Citation11].
In the present study, molecular analysis (RAPD and ISSR) was applied beside biochemical analysis (SDS-PAGE) to assess genetic variation within and among Egyptian Mesembryanthemum species growing naturally in different coastal regions of Egypt.
2 Materials and methods
2.1 Population sampling
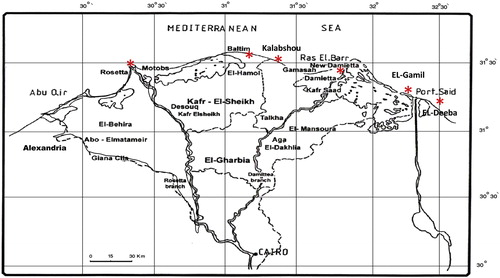
Plant samples were collected from twelve natural populations of three Mesembryanthemum species representing different localities along the International Coastal Road as listed in and represented in . M. crystallinum L. and M. nodiflorum L. populations were collected from five localities (Port said, Demiatta, El-Dakahlia, Kafr El-sheikh and El-Beheira), while M. forsskaolii Hochst. ex Bioss. populations were collected from two localities only (El-Gamil and Port said city). The number of sampled populations depended on the actual population size. Seeds and fresh young leaves were collected as bulked samples from each population of each species in the study area for genetic analyses.
Table 1 Location and habitat type of the sampled twelve populations representing the three studied species of Mesembryanthemum L.
2.2 Biochemical analysis (SDS-PAGE)
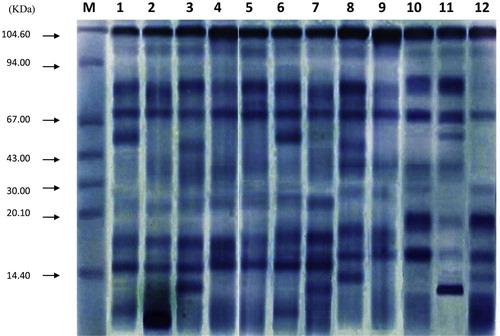
Sodium dedocylsulfate polyacrylamide gel electrophorosis (SDS-PAGE) as described by Laemmli [Citation16] was used to differentiate between twelve populations of studied M. crystallinum, M. forsskaolii and M. nodiflorum seeds by their protein profiles. It is the standard method for separating and differentiating protein according to their molecular weights. The banding profile in the gel was photographed and the number of bands for each taxon was scored.
2.3 Plant material and DNA extraction
Genomic DNA was extracted using a modified protocol of Dellaporta [Citation17]. The amount of DNA was quantified by agarose gel electrophoresis. DNA samples were stored at −20 °C for RAPD and ISSR analyses.
2.4 RAPD amplifications
Polymerase chain reaction (PCR) was conducted according to Williams et al. [Citation13]. Twelve RAPD primers were initially screened but only five primers (OPO9, OPO10, OPO11, OPO12 and OPO13) were used in the analysis and listed in . The generated clear and reproducible banding patterns were used to evaluate genetic diversity. The reaction conditions were optimized and mixtures were composed of dNTPs (5 μl), MgCl2 (5 μl), 10× buffer, primer (5 μl), DNA (10 μl), promega Taq DNA polymerase (2.5 units). DNA amplification was carried out in a thermocycler (PTC-100) programmed for 95 °C for 5 min (one cycle); followed by 94 °C for 1 min, 36 °C for 1 min and 72 °C for 2 min (45 cycle); 72 °C for 5 min (one cycle), then 4 °C (infinitive). Amplification products (10 μl) were mixed with 5 μl bromophenol blue separated on 1.5% agarose gel and stained with 3 μl ethidium bromide then photographed.
2.5 ISSR amplifications
ISSR-PCR reactions were conducted according to Zietkiewicz et al. [Citation18] using five selective ISSR primers for the genotypes as (HB11, HB12, HB13, HB14 and HB15) which were synthesized on an ABI 392 DNA/RNA synthesizer (Applied Biosystems) at AGERI, ARC (Egypt) and represented in . DNA amplification was carried out in Stratgene PCR 96 which was programmed for denaturation (one cycle) at 94 °C for 2 min, followed by 30 cycles: as follows 94 °C for 30 s, 44 °C for 45 s, 72 °C for 1 min, and finally one cycle extension at 72 °C for 20 min, and 4 °C (infinitive). Fifteen μl of PCR-products were resolved in 1.5% GTG agarose gel electrophoresis with 1× TAE running buffer. The run was performed at 80 V for 180 min and the gel was stained with ethidium bromide then photographed.
The gels of DNA and protein were visualized and photographed by gel documentation system (Biometra Bio Doc Analyze 2000) under UV transilluminator at central laboratory of Desert Research Center. The bands were scored as present (1) or absent (0) regardless of their intensity. Only the clear and reproducible bands were recorded. The smeared and weak ones were excluded from analysis. The binary data generated were used to estimate levels of polymorphism by dividing the polymorphic bands by the total number of scored bands.
2.6 Genetic diversity and structure
Using GenAlEX 6.2 [Citation19], standard measures of genetic diversity for each species were calculated including the number of different alleles (Na), the number of effective alleles (Ne) = 1/(p2 + q2), Shannon's information index (SI) = −1(p Ln (p) + q Ln(q)), the expected heterozygosity (He) = 2pq, the unbiased expected heterozygosity (UHe) = (2N/(2N−1)) He, and percentage of polymorphic loci (%P). All the measurements were assessed and represented as mean and standard error (SE) over loci within populations and over loci among populations within the species [Citation20]. The genetic differences (PhiPT) among species were estimated using the Analysis of Molecular Variance (AMOVA) procedure [Citation21] to investigate the hierarchical partitioning of genetic variation among species. PhiPT was calculated as the proportion of the variance among species, relative to the total variance. Unbiased genetic identity and distance [Citation23] were calculated between pairs of species using GenAlEX 6.2 [Citation22].
3 Results
3.1 SDS-PAGE electrophoretic pattern
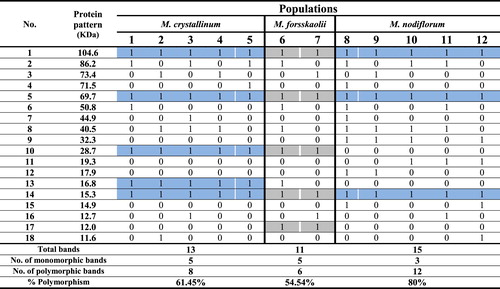
The electrophoretic analysis of protein samples showed a distinctive electrophoretic pattern. The electrophoretic protein profile of Mesembryanthemum species represented in18 bands with molecular weight ranged from 11.6 to 104.61 (KDa) as shown in and illustrated in . In M. crystallinum, the protein profile consisted of 13 bands; 5 bands were recorded as monomorphic and 8 bands were polymorphic. Five unique bands were found of molecular weights as follow (71.5, 50.8, 44.9, 12.7 and 11.6 KDa). The percentage of polymorphism was calculated as 61.54%. In M. forsskaolii, a total of 11 bands were revealed in which 5 bands were monomorphic and 6 bands were polymorphic. Six unique bands were recorded with molecular weight (86.2, 73.4, 50.8, 40.5, 16.8 and 12.7 KDa). The percentage of polymorphism was 54.54%. In M. nodiflorum, 15 bands were found in which 3 bands were monomorphic and 12 bands were polymorphic. Four unique bands were scored with molecular weight (73.4, 44.9, 12.7 and 11.6 KDa). The percentage of polymorphism was estimated as 80%.
3.2 Molecular analysis
The result of DNA profile using RAPD analysis for 5 primers (OPO9, OPO10, OPO11, OPO12 and OPO13) and ISSR analysis for 5 primers (HB11, HB12, HB13, HB14, and HB15) are shown in . The range of bands products numbers of monomorphic and polymorphic bands produced by different primers are shown in .
Table 3 List of primers and their nucleotide sequences, range of band products, number of common and polymorphic bands and the percentage of polymorphism produced by different RAPD and ISSR primers.
Table 4 RAPD and ISSR profiles of different genotypes in Mesembryanthemum L. using five primers for each marker, for population number see .
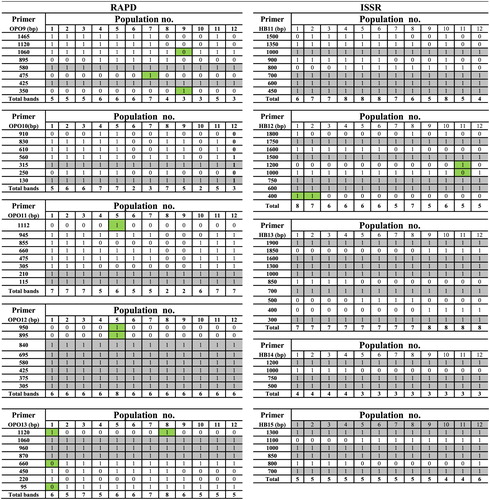
Table 5 Comparative analysis of bands characteristics generated by RAPD and ISSR markers in Mesembryanthemum species.
Plate 2 DNA polymorphasim based on RAPD-PCR and ISSR-PCR analysis of Mesembryanthemum L. populations in Egypt, for population number see .
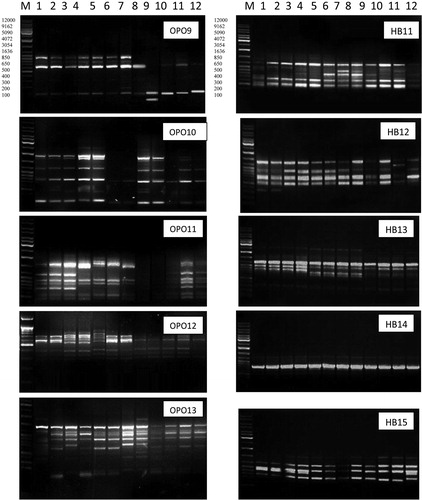
3.2.1 RAPD analysis
RAPD-PCR profile showed a high polymorphism in Mesembryanthemum populations. A total of 39 reproducible and clear bands were amplified with different lengths, ranging from 95 to 1465 bp in which 24 bands were polymorphic. Number of polymorphic bands varied between 2 and 6. The average number of polymorphic fragments per primer was 4.8. The percentage of polymorphism ranged from minimum value of 25% in (OPO12) to maximum value of 75% in (OPO9 and OPO11) with an average of 61.78% as shown in and .
With primer OPO12, there were two characteristic unique bands of molecular weights 950 bp and 895 bp for M. crystallinum. Moreover, primer Opo13 band of molecular weight 1120 bp can be considered as unique (private allele) for M. crystallinum. The results also revealed that, primer OPO9, OPO10 and OPO13 bands of molecular weights 475 bp, 560 bp, and 220 bp respectively, are specific (private alleles) for M. forsskaolii. While primer OPO9 bands with molecular weights 350 bp, 1465 bp, and 1120 bp present in one site and absent in the others. Also, primer OPO10 bands of molecular weights 250 bp and 910 bp present in one site and absent in the others. In primer OPO13, the unique bands of molecular weight 450 bp and 1120 bp were specific for M. nodiflorum.
3.2.2 ISSR analysis
Concerning ISSR analysis, the primers yielded 37 highly reproducible ISSR bands ranging from 300 to 1900 bp in size. As shown in and , 16 of the bands were polymorphic with an average of 3.2 polymorphic bands per primer, while the remaining 21 bands were monomorphic. The percentage of polymorphism was found in the range of minimum value of 25% (HB14) to maximum value of 55.5% (HB12), with an average of 40.77%.
The results indicated that primers HB11 and HB12 bands of molecular weights 900 bp and 1600 bp respectively, produced specific (private alleles) bands for M. forsskaolii in population (6). It was found that bands of the molecular weights 1800 bp, 500 bp, and 1100 bp corresponding to the primers HB12, HB13, and HB15, respectively, are unique bands (private alleles) for M. crystallinum. In M. nodiflorum, the bands of molecular weights 900 bp and 1500 bp belonging to the primer HB11were present in only one population (10) and absent from the rest of sites populations. Also, bands of molecular weights1200 bp and 1600 bp belonging to primer HB12 and bands of molecular weights 500 bp and 800 bp belonging to primers HB13 and HB15, respectively, were recorded as unique bands (i.e. private alleles recorded in only one population within the species) for M. nodiflorum in ().
3.3 Genetic diversity and structure
Although using RAPD and ISSR genetic markers resulted in higher number of bands than SDS-PAGE (39, 37 and 18 bands, respectively), all measured diversity parameters were higher using SDS-PAGE than RAPD followed by ISSR, e.g. the average of heteromorphism all over the studied three species was 48.15% using SDS-PAGE, while it was only 29.06% using RAPD and 20.72% using ISSR (). RAPD and ISSR markers agreed on assigning higher diversity values to M. nodiflorum followed by M. crystallinum than M. forsskaolii. On other hand, SDS-PAGE assigned M. crystallinum the highest diversity values followed by M. nodiflorum. It assigned M. forsskaolii the lowest diversity values (). Results should be treated with caution given the sample size in the M. forsskaolii is less than 5.
Table 6 Genetic diversity parameters revealed by different genetic markers (RAPD, ISSR and SDS-PAGE).
Generally, AMOVA results revealed that proportion of total genetic variation found among species (PhiPT) was high in (). Relatively, it was lower for RAPD maker (23%) than for SDS-PAGE marker (27%). ISSR resulted in a fairy high total genetic variation among species (47%). Also, the percentages of molecular variance within species populations were recorded as 73%, 77% and 53% using SDS-PAGE, RAPD and ISSR markers respectively.
4 Discussion
Assessing genetic diversity is an important precursor in any population genetic investigation that it affects the evolutionary potential of a species and populations [Citation23]. This study offered some detailed analysis of the genetic variation and structure for three Egyptian Mesembryanthemum species growing in different natural habitat based on biochemical and molecular markers that have been proven to be valuable for the determination of genetic diversity.
RAPD primers OPO11 bands with molecular weights 1112 bp and 950 bp and OPO12 bands with molecular weight 895 bp were private alleles distinctive for M. crystallinum populations at Rosetta city (population 5). Band with molecular weight of 475 bp resulted out using primer OPO9 is a private allele for M. forsskaolii population at Port said city (population 7) and band with molecular weight of 350 bp of primer OPO9 is a private allele for M. nodiflorum population at New Demiatta city (population 9). Meanwhile, 1200 bp band resulted from using HB12 primer in ISSR marker is a private allele for M. nodiflorum population at Baltim city (population 11).
The results indicated that molecular analysis by using RAPD and ISSR techniques may be good tools for DNA fingerprinting, detecting the genetic variations and identifying of Mesembryanthemum species collected from different locations. Variation in DNA sequences lead to polymorphism. Greater polymorphism reflects higher genetic diversity. Polymorphism percentages of different used markers (RAPD, ISSR and SDS-PAGE) were recorded as follow 61.78%, 40, 77% and 80% respectively.
As reported in previous studies [Citation24] on the genetic diversity among the three cogeneric wild species (M. crystallinum L., M. nodiflorum L. and M. forsskaolii Hochst. ex Bioss.) and the relationships between these species were determined by using RAPD and isozymes, the results showed that a notable polymorphic bands in ten RAPD markers giving a little variation in the genetic divergence between studied species. Similar results were reported in assessment of genetic diversity of Pancratium L. and Launaea Cass. species by using RAPD and ISSR markers [Citation25,Citation26].
Random amplified polymorphic DNA (RAPD) and inter simple sequence repeats (ISSR) markers require only small amounts of DNA without involving radioactive labels and are simpler as well as faster. RAPD has proven to be quite efficient in detecting genetic variations and used for genetic diversity and for identifying germplasm of plant species [Citation27,Citation28]. ISSR has been shown to provide a powerful, rapid, simple, reproducible and inexpensive means to assess genetic diversity and identify differences between closely related cultivars in many species [Citation29]. ISSR may be a valuable tool for DNA fingerprinting of plant cultivars [Citation30]. Yildiz et al. [Citation31], stated that the genetic variation as measured by ISSR, SRAP and RAPD markers also revealed high diversity among Turkish melon genotypes (H = 0.30, I = 0.45 and 87.6%).
Comparing with other short-lived, outcrossing perennial species (P = 43.7% and 32.1, He = 0.123) [Citation32], M. crystallinum and M. nodiflorum exhibited within range genetic diversity. RAPD and ISSR markers exhibited lower genetic diversity for M. forsskaolii. This may be attributed to sample size which is less than 5. On the local-scale, three Ballota species (Lamiaceae); B. undulata (Fresen.) Benth., B. saxatilis C. Presel, and B. kaiseri Täckh. native to southern Sinai have been analyzed for allozyme diversity [Citation33] and Hypericum sinaicum and Origanum syriacum subspecies sinaicum have been analyzed for AFLPs [Citation34]. Comparing with these species which are perennial outcrossing herbs M. crystallinum and M. nodiflorum had lower genetic diversity. For comparisons across studies using different genetic markers, estimates of heterozygosity appear to be the most suitable and commonly reported parameters, although these are influenced by choice of bands and loci [Citation35].
Notes
Peer review under responsibility of Mansoura University.
References
- C.KlakP.V.BruynsT.A.J.HeddersonA phylogeny and new classification for Mesembryanthemoideae (Aizoaceae)Taxon562007737756
- L.BoulosFlora of Egypt1999Al-Hadara PublishingCairo, Egypt
- F.M.El-ShayebH.El-TantawyA.El-KholiStudies on the distribution of wild Mesembryanthemum species growing in EgyptJ Union Arab Biologists Cairo91999253268
- H.FallehR.KsouriF.MediniS.GuyotC.AbdellyC.MangeAntioxidant activity and phenolic composition of the medicinal and edible halophyte Mesembryanthemum edule L.Ind Crop Prod34201110661071
- S.M.Abdel RahmanS.A.Abd-EllatifS.F.DerazA.A.KhalilAntibacterial activity of some wild medicinal plants collected from western Mediterranean coast, Egypt: natural alternatives for infectious disease treatmentAfr J Biotech10201110733 e43
- B.IbtissemC.AbdellyS.SfarAntioxidant and antibacterial properties of Mesembryanthemum crystallinum and Carpobrotus edulis extractsAdv Chem Eng Sci22012359 e65
- Bohnert LaboratoriesMesembryanthemum2003 Available at:http://www.life.uiuc.edu/bohnert/projects/mesem.html
- A.I.MustafaM.S.Al-JassirM.A.NawawyS.E.AhmedStudies on samh seeds (Mesembryanthemum forsskalei Hochst) growing in Saudi Arabia: 3. Utilization of samh seeds in bakery productsPlant foods Hum Nutr481995279286
- M.ChaiebM.BoukhrisFlore succincte et illustrée des zones arides et sahariennes de Tunisie1998Association pour la protection de la nature et de l’environnementSfax Tu
- Q.ZhangA.SaghamA.KleinhofsComparative diversity analysis of RFLPs and isozymes within and among populations of Hordeum vulgare ssp. spontaneumGenetics1341993909916
- L.MondiniA.NooraniM.A.PagnottaAssessing plant genetic diversity by molecular toolsDiversity20091935 doi:3390/d1010019
- A.KarpO.SebergM.BuiattiMolecular techniques in the assessment of botanical diversityAnn Bot781996143149
- J.G.K.WilliamsA.R.KubelikK.J.LivakJ.A.RafaskiS.V.D.N.A.TingeyPolymorphisms amplified by arbitrary primers are useful as genetic markersNucl Acids Res18199065316535
- M.P.ReddyN.SarlaE.A.SiddiqInter simple sequence repeat (ISSR) polymorphism and its application in plant breedingEuphytica1282002917 10.1023/A: 1022205000732
- T.M.CulleyA.D.WOLFEPopulation genetic structure of the cleistogamous plant species Viola pubescens Aiton (Violaceae), as indicated by allozyme and ISSR markersHeredity86200154555610.1046/j.1365-2540.2001.00875.x
- U.K.LaemmliCleavage of structural proteins during the assembly of the head of bacteriophage T4Nature2271970680685
- S.L.DellaportaJ.WoodJ.B.HicksA plant DNA minipreperation: version IIPlant Mol Biol Rep119831921
- E.ZietkiewiczA.RafalskiD.LabudaGenome finger-printing by simple sequence repeat (SSR) –anchored polymerase chain reaction amplificationGenomics201994176183
- R.PeakallP.E.SmouseGENALEX 6: genetic analysis in Excel. Population genetic software for teaching and researchMol Ecol Notes62006288295http://dx.doi.org/10.1111/j.1471-8286.2005.01155.x
- M.NeiAnalysis of gene diversity in subdivided populationsProceedings of the National Academy of Sciences, USA70197333213323
- L.ExcoffierP.E.SmouseJ.M.QuattroAnalysis of molecular variance inferred from metric distances among DNA haplotypes : application to human mitochondrial DNA restriction dataGenetics1992131473
- M.NeiGenetic distance between populationsAm Nat1061972283292
- X.MaX.Q.ZhangY.H.ZhouS.Q.BaiW.LiuAssessing genetic diversity of Elymus sibiricus (Poaceae: Triticeae ) population from Qinghai – Tibet Plateau by ISSR markersBiochem Syst Eco362008514522
- A.M.FawzyRAPD and isozyme studies of Mesembryanthemum species in Egyptian floraMinia J Agric Res Dev27520079871005
- M.I.SolimanR.M.RizkF.M.El-SaiedN.M.El-ErakyGenetic variation of some Pancratium L. species in Egypt related to accessions environment and geographical isolationBiol Chem Environ Sci7120126180
- M.I.SolimanF.M.ElsaiedL.Z.SamaanG.T.GhoniemGenetic diversity of some Launaea Cass. species EgyptJ. Environ Sci Mans Uni43120147996
- J.WelshM.McClellandFingerprinting genomes using PCR with arbitrary primersNucleic Acids Res18199072137218
- J.KapteynJ.E.SimonThe use of RAPDs for assessment of identity, diversity and quality of EchinaceaJ.JanickA.WhipkeyTrends in new crops and new uses2002ASHS Press Alexandria VA 509-13
- A.GonzalezA.CoulsonR.BrettellDevelopment of DNA markers (ISSRs) in mangoActa Hortic5752002139143
- A.ShiS.KanatartziM.MmbagaP.ChenDevelopment of ISSR PCR markers for diversity study in dogwood (Cormus spp.)Agric Biol J N Am132010189194
- M.YildizE.EkbicD.KelesS.SensoyK.AbakUse of ISSR, SRAP and RAPD markers to assess genetic diversity in Turkish melonsSci Hortic1302011349353
- J.L.HamrickM.J.W.GodtAllozyme diversity in plant speciesA.H.D.BrownM.T.CleggA.L.KahlerB.S.WeirPlant population genetics, breeding, and genetic resources198943
- M.S.ZaghloulJ.L.HamrickA.A.MoustafaW.M.KamelR.El-GhareebGenetic diversity within and among Sinai populations of three Ballota species (Lamiaceae)J Hered45200697http://dx.doi.org/10.1093/jhered/esj008
- M.S.ZaghloulP.PoschlodC.ReischGenetic variation in Sinai's range-restricted plant taxa Hypericum sinaicum and Origanum syriacum subsp. sinaicum and its conservational implicationsPlant Eco Evol1472014187201
- H.NybomComparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plantsMol Ecol13200411421155http://dx.doi.org/10.1111/j.1365-294X.2004.02141.x