Abstract
Ecdysteroid signal transduction plays a major role in insect metamorphosis, 20-hydroxyecdysone (20E) binds to the nuclear receptor composed of the ecdysone receptor ligand binding domine (EcR-LBD) and triggers the developmental transitions. Ariadne merione ecdysone receptor (AmEcR) cDNA was amplified and partially sequenced of about 553 bp, which encodes a polypeptide of 184 amino acids (aa). The theoretical molecular weight (MW), isoelectric point (pI) and aliphatic index of the deduced AmEcR protein were predicted using BIOEDIT (v7.2.5) to be 21.192 kDa, 9.31 and 101.739 respectively. Identified ecdysone receptor gene of A. merione showed maximum similarity with Precis coenia gene. In this research, we have employed ligand-receptor engineering technique to screen a specific compound which plays antagonist role and assist to formulate an insect specific pesticide. The EcR protein 3D structure of AmEcR modeled using Schrödinger maestro and virtual screening was performed using 5554 molecules from Zinc database, where ZINC20031812 showed highest glide score of −6.257 and Etoxazole chosen on literature basis and showed best glide score −6.671. We have compared the antagonist with agonist (20E) by molecular dynamics (MD) simulation. Root Mean Square Deviation (RMSD) value of agonist and antagonist indicates the binding were stable in water with a range of distance from 2.3 to 2.6 Å, 1.8 to 2.3 Å and 1.9 to 2.3 Å with a variation over the time scale of 1 ps. Since Etoxazole and ZINC20031812 are antagonists, computationally they were more stable than 20E.
1 Introduction
A common feature between both types of hemimetabolous and holometabolous insect development is that the periodic pulses of the steroid hormone 20 hydroxyecdysone (20E) which dictate each developmental transition. The ecdysone receptor complex is the key element, which enacts the ecdysteroid-induced physiological and morphological changes during insect moulting regulated by ecdysteroid hormones like 20-hydroxyecdysone (20E) and its analogs that bind to the ligand-binding domain of the ecdysone receptor [Citation1]. Although the molecular action of 20E has been extensively studied in holometabolous insects and the data on hemimetabolous is scarce [Citation2]. The hormone 20E is lipophilic in nature, which synthesized and released into the hemolymph, and enters into the cells are responsible for metamorphosis. The Ecdysone receptor gene is a nuclear receptor with 2 major domain named as ligand binding domain (LBD) and DNA binding domain (DBD), it is seen in cytoplasm at inactive state. The 20E binds to LBD of ecdysone receptor gene and it activates the DBD. The basic structure of ecdysone receptor protein (EcR) consists of five domains referred to as A/B-transcriptional activation domain, C-DNA-binding domain, D-hinge region and E-ligand-binding domain [Citation3].
The moulting process is initiated by a number of transcription factors in the nuclear receptor super family. This result showed in the up-regulation of several late genes in the hormone pathway and help in mediating the moulting process [Citation4]. These nuclear receptors have ability to travel through the nuclear pores due to its heterodimeric nature, and bind to specific sites of DNA, which undergoes active replication, chromosome remodeling and finally results in translation. Cuticle degradation and formation of new cuticle are one of the major signaling functions of ecdysone during moulting and N-acetyl-β-d-glucosamine (chitin) is a major component of the insect cuticle [Citation5].
Some of the members belonging to lepidopteran order are serious agriculture pests, which destroys the crops by defoliating. The present study is hypothesized based on the bioinformatics analysis of specific developmental gene and protein encoded by the same. The targeting of specific developmental protein (insect specific) may help in controlling the target pests. The screened compounds mimic 20 hydroxyecdysone which precisely interfere with receptor functions and helps to control the mass number of agriculture pests. This could be achieved by using an effective bioinformatics tools to design a perfect molecule to dock with ecdysone receptor proteins.
2 Materials and methods
2.1 RNA isolation
Total RNA was isolated from fourth instar of A. merione (at inter-moult stage) using TRIzol Reagent (Ambion®, Life Technology, USA). One individual (=100 mg) of A. merione fourth instar was ground well with 1 ml of TRIzol reagent using sterile Teflon hand homogenizer. The tubes containing the homogenate were incubated for 5 min at RT to permit complete dissociation of the nucleoprotein complex, 200 μl of pre-chilled chloroform was added, mixed vigorously for 15 s and the suspension was incubated at RT for 2–3 min followed by centrifugation (7500×g, 4 °C, 15 min). The supernatant (∼500 μl) was transferred to a sterile tube, after which 500 μl of 75% isopropanol was added and incubated at RT for 10 min, followed by centrifugation (7500×g, 4 °C, 15 min). The supernatant was discarded, then 1 ml of 75% ethanol was added to the pellet, vortexed and centrifuged (6800×g, 4 °C, 5 min). The supernatant was completely discarded, finally the pellet was air-dried, re-suspended in 20 μl of DEPC water and stored at −80 °C for further use.
2.2 Polymerase chain reaction
The isolated total RNA from A. merione larvae was reverse transcribed according to the manufacturer's protocol using first strand cDNA Synthesis Kit (Roche, Germany). Two gene specific primers, forward primer (5′-AGATGACCATCCTCACCGTG-3′) and reverse primer (5′-ACGTCCCAGATCTCCTCGAG-3′) were designed based on the conserved region of gene sequences in the other lepidopterans EcRs. The annealing temperature was fixed at 57 °C for 35 cycles. PCR products were purified by QIAquick gel extraction kit (Qiagen).
2.3 DNA sequencing and sequence analysis
The purified and extracted sample was sequenced at Eurofins Scientific Ltd. (Bangalore, India) using Sanger dideoxy technology. The obtained sequence was analyzed using BIOEDIT software (ver.7.2.5) and alignment was done for AmEcR nucleotide and its encoding protein.
2.4 Homology modeling and structural validation
The method to predict ab initio modeling for AmEcR translated protein sequence by using the Schrödinger maestro (ver.9.3) (Schrödinger Inc.). The Prime-SP (ver.3.1) (Standard Precision) is used and it facilitates the comparative modeling that includes alignment, build structure, fold recognition and molecular mechanics-generalized born model augmented with the hydrophobic solvent accessible surface area calculations. Modeled structure was validated for its property in the range and structural similarity from sequence using ProCheck. The residues of Glycine and Proline were analyzed against total number of amino acids present in AmEcR modeled structure. This AmEcR modeled structure was subjected to refinement and validation using Ramachandran plot.
2.5 Grid generation
Glide program was employed to set up the Receptor Grid Generation by clicking the Receptor Grid Generation Panel. Default parameters were used and no special constraints were incorporated during grid generation. Default grid size was adopted for all the active sites.
2.6 Ligand retrieval
Acetamiprid, Chromafenozide, Dibenzoylhydrazines, Etoxazole, Fenpyroximate, Methoxyfenozide, Pyriproxyfen and Tebufenozide were selected for the study on the basis of literature survey [Citation6,Citation7] , retrieved from PubChem database. On the other hand, structure data file (SDF) was downloaded from ZINC database by setting the compounds property based on Tice rule. Tice rule states that the potential insecticidal compounds should have: (1) molecular weight less ≤ 500 g/mol, (2) number of hydrogen donors ≤ 3, (3) number of hydrogen bond acceptors ≤ 12, (4) log P partition co-efficient (lipophilicity) ≤ 5, and (5) number of rotatable bonds ≤ 12 [Citation7,Citation8] . A single SDF file with 29 Mega byte was downloaded, which contained 5554 compounds. 20 Hydroxyecdysone (20E) was retrieved from PubChem database and served as control for comparison study.
2.7 Virtual screening and docking
Virtual screening was performed in Schrödinger module using SDF file of 5554 compounds from Zinc data base. This SDF file was imported into the system. Virtual screening was initiated, then 500 iteration was given to analyze the ligand structure and ability to bind in the receptor. Based on virtual screening the top scored, single ligand was utilized for docking studies. The Virtual Screening workflow panel sets up the input files for LigPrep (ver.2.6), QikProp (ver.3.5), and Glide (ver.5.8) ligand docking and submits them to the selected host in order. Glide results were examined with an emphasis on visual rather than numerical appraisal. The first set of exercises used the Project Table to display the results of the Standard Precision (SP) Glide docking job, examined individual ligand poses and their contacts with the input receptor structure. The second set of exercises used the Glide express precision (XP) visualizer panel to display information on the terms in the Glide XP scoring function that contribute to the ligand binding.
2.8 Methodology and parameterization for molecular simulation of protein and ligand complex
The molecular simulations were performed using Desmond (ver.3.1) (D.E. Shaw Research, New York) with an inbuilt Optimized Potential Ligand Simulation (OPLS-2005), which provides a strong framework for the calculation of bonding energies between the biomolecular structures. In the current study, molecular dynamics was performed to detect the stability of AmEcR – 20E complex, AmEcR – Etoxazole and AmEcR – ZINC20031812 complex in water solvent. After 1000 ps equilibration, a trajectory for 1000 ps was generated for 100 samples. The Root Mean Square Deviation (RMSD) is used to measure the scalar distance between atoms of the same type for two structures. In the current study, we used the RMSD between C (alpha) atoms to measure the fit between protein homologs. In this calculation, we used the RMSD of heavy-atoms to compare the spatial deviation between structures in time and the original structure (at time = 0 ps). Typically, the RMSD on heavy-atoms should not change more than 3 Å within a nanosecond of molecular simulation time.
3 Results and discussion
3.1 DNA sequencing and sequence analysis
On subsequent gene cleaning and sequencing of the PCR product, nucleic acid content using BIOEDIT (ver.7.2.5) revealed the presence of 32.91% cytosine followed by 26.58, 21.70 and 18.81% guanine, adenine and thymine respectively. Based on these numbers of nucleotides, the length of the partial Coding sequence region (CDS) of EcR possessed 553 bp and alignment was done for AmEcR nucleotide and its encoding protein (). AmEcR gene sequences were submitted in NCBI database (Accession number KJ652504).
3.2 Homology modeling and structural validation
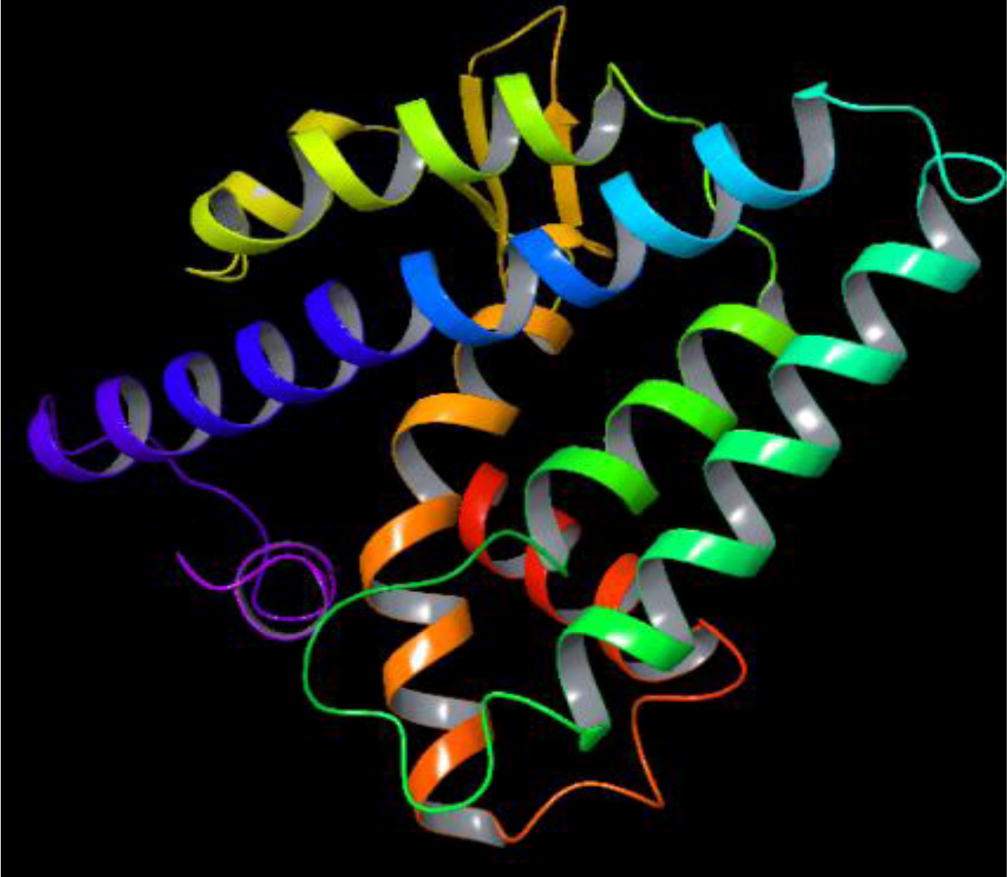
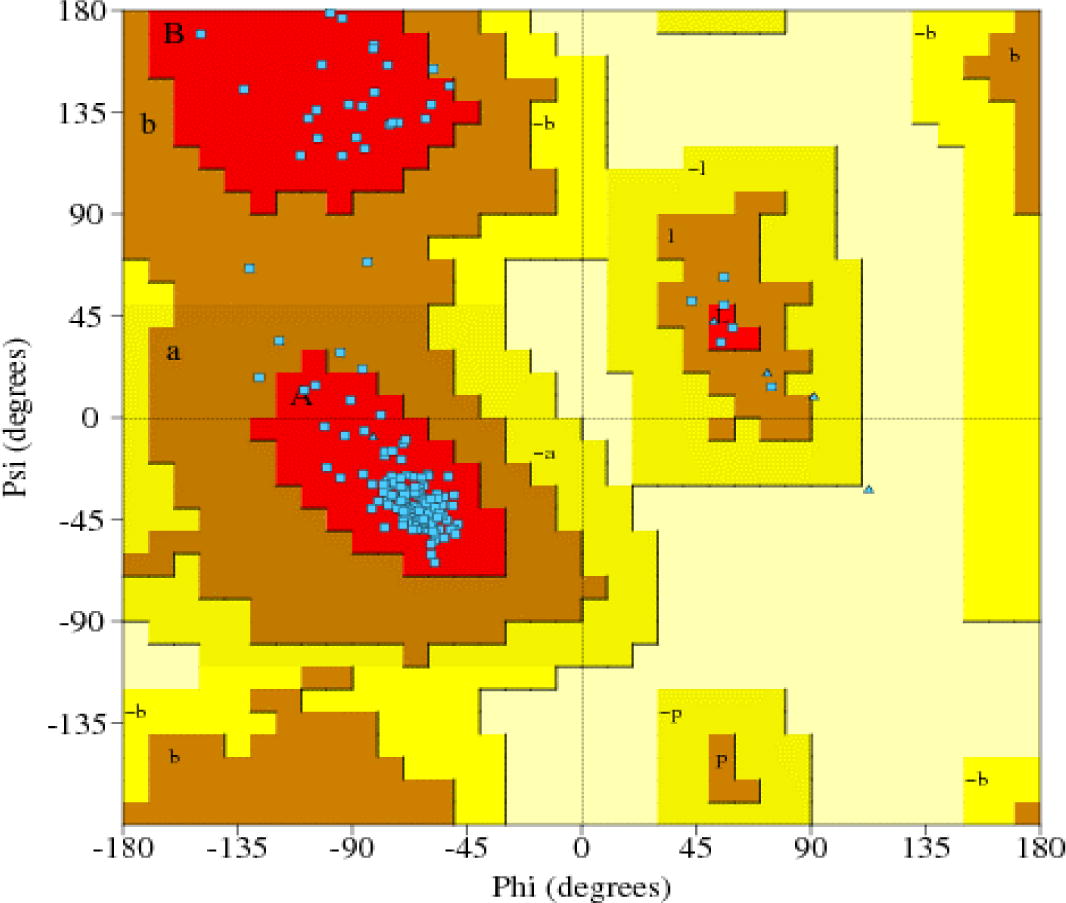
AmEcR sequence was subjected to perform homology modeling against Protein Databank which has the structural conformation of each atom’s configuration. Schrödinger (ver.9.3) module chosed the ECR-LBD protein structure of Heliothis virescens (PDB accession code – 1R1K_D) sequence as a template to construct the AmEcR protein structure (). This AmEcR exhibited 89.31% similarity with template protein. Modeled structure of AmEcR was validated using ProCheck. An ideally, the structure showed about 86% of the residues in their core region, which is about 94% similar to that of the template structure in the alignment procedure. The percentage of residues in the core regions is the better guide to stereo-chemical quality (; ).
Table 1 Percent residues of AmEcR localized in Ramachandran Plot.
3.3 Screening of compounds
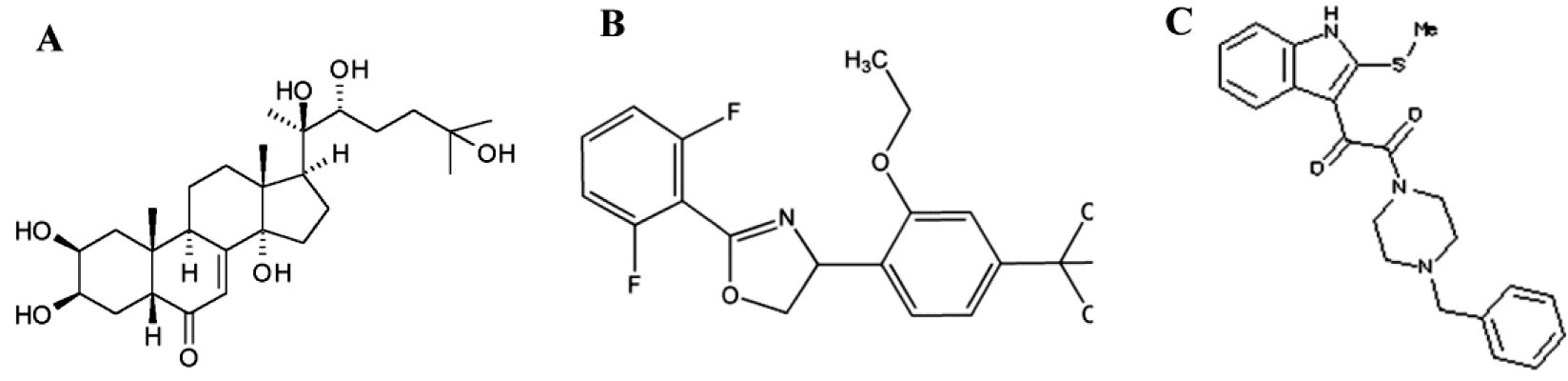
The known 20E ligand was involved during the metamorphosis for binding to the receptor. Subsequently, 5554 compounds were screened out of which two compounds namely Etoxazole and ZINC20031812 showed docking compatibility and hence utilized for further studies (, ). The molecules have been assigned biologically relevant protonation states and are annotated with properties such as molecular weight, calculated Log P, and number of rotatable bonds. This database is available for free download (http://zinc.docking.org) in several common file formats including SMILES, mol2, 3D SDF, and DOCK flexibase format [Citation9]. Virtual screening helps to identify novel non-steroidal ligand that are similar to the known EcR ligand [Citation10].
Table 2 The molecular property of selected agonist and antagonist molecule for docking studies based upon Tice rule.
3.4 Docking and protein-ligand interaction profile
The docking of 20E to the receptor model revealed that the ligand molecule can interact with the receptor in a similar manner to other steroid hormone-receptor complexes [Citation11]. In the current study, in silico docking shows the binding domain in protein and interaction of ligands in particular pockets were shown in the . Tyrosine in the 64th position was common binding amino acid with all ligands that shows these antagonists binds in the same pocket where the authentic 20E binds. After analysis of several crystal structures, it revealed that the DNA-binding domain (DBD) and ligand binding domain (LBD) are highly conserved in insect EcRs [Citation12]. An approach was initiated by using the crystal structure of the ligand-binding domain of the ecdysone receptor (EcR) of the moth Heliotis virescens as well as virtual molecule libraries of analogues of known diacyl-hydrazine (DAH) type ecdysteroid agonists. By docking DAH with binding pocket of EcR followed by CoMFA (Comparative Molecular Field Analysis) and CoMSIA (Comparative Molecular Similarity Indices Analysis) of the docked conformations, hitherto unexplored regions of the receptor cavity could be mapped [Citation13].
Table 3 AmEcR modeled protein and ligands interaction profile.
3.5 Modeled AmEcR vs 20E
Molecular docking between AmEcR with 20E showed the ligand interacted at 5 sites of AmEcR having residual atom types ALA 54 (H), THR 2 (H), TYR 64 (H), ASN 160 (O), ARG 39 (H) receptor via C (Carbon), H (Hydrogen) and O (Oxygen) forming carbon, hydrogen and oxygen bonds with the bond distance of 2.02, 2.37, 2.08, 1.81, 2.33 and 2.13 Å (A). The higher interaction of AmEcR with 20E can be noticed from the Glide score −8.248 (). The hydrogen bonding is one of the important physicochemical properties for ligand binding to the ecdysteroid receptor, the number of possible hydrogen bonding between the ligand molecule and the receptor was manually counted in the modeled ligand–receptor complex [Citation10]. During the metamorphosis, the LBD plays major roles, acts as a receptor dimerization, ligand recognition and cofactor interactions [Citation14]. Ecdysone receptor and ligand binding models attempt to explain how 20E and dibenzoylhydrazines interact with the ligand-binding pocket and homology model complexes offers new insights that can be exploited in the rational design of new environmentally safe insecticides [Citation11Citation[12]Citation[13]Citation[14]–Citation15] .
Fig. 5 Interaction profile of AmEcR with the tested ligands, A) AmEcR vs 20 hydroxyecdysone (20E), B) AmEcR vs Etoxazole and C) AmEcR vs ZINC20031812. Tyrosine in 64th position was a common binding amino acid with all ligands that shows the Etoxazole and ZINC20031812 (antagonist) binds in the same pocket where the 20E binds.
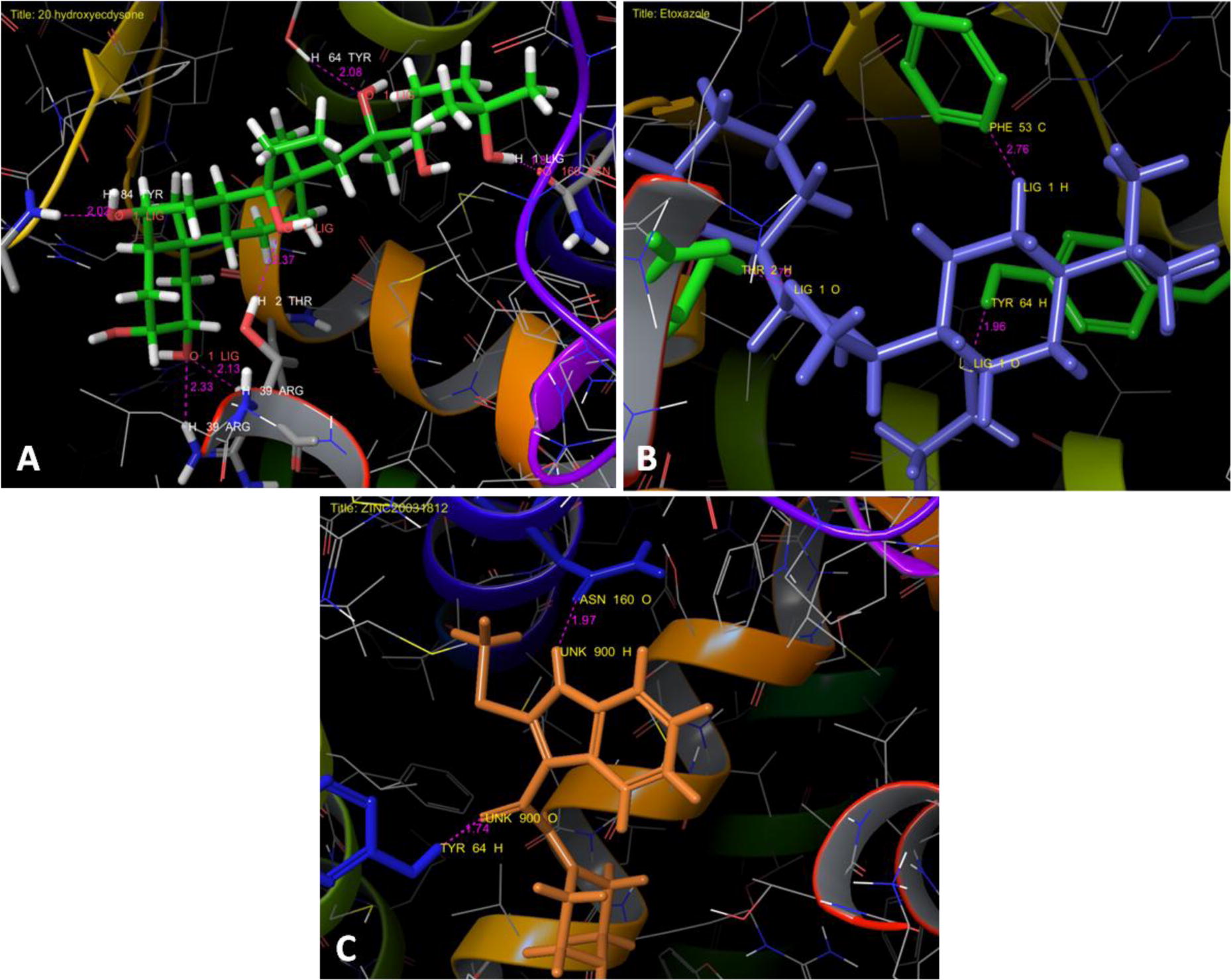
3.6 Modeled AmEcR vs Etoxazole
Interaction of AmEcR with Etoxazole showed that this ligand interacted at 3 sites of the AmEcR having residual atom types PHE 53 (C), TYR 64(H) and THR 2(H) with the binding distance of 2.70,1.96 and1.75 Å (B). The Glide score −6.671 indicated the highest interaction of AmEcR with Etoxazole (). The LBD contains the ligand-binding pocket (LBP), which binds ecdysteroids as well as certain nonsteroidal EcR agonists such as the DAH-based insecticides [Citation16].
3.7 Modeled AmEcR vs ZINC20031812
Docking ZINC20031812 with AmEcR model showed that the ligand interacted at 2 sites of the AmEcR having residual atom types ASN 160 (O), THR 64 (H) and its binding distance are 1.97 and 1.74 Å (C). The Glide score of −6.257 indicated the potential interaction of AmEcR with ZINC20031812 ().
3.8 Molecular simulation of AmEcR with agonist and antagonist complexes
Molecular simulation, the positions and velocities of particles corresponding to atoms evolve according to the laws of classical physics [Citation17]. In this paper, we showed the super-imposed conformational structure for AmEcR-20E, AmEcR-Etoxazole and AmEcR-ZINC20031812 complex by calculating RMSD value. RMSD measures the average change in displacement of selected atoms for a specified frame with comparison to a reference frame, which is intended for all frames in the trajectory. In protein RMSD, protein frames would be aligned initially on the reference frame backbone where the RMSD is calculated based on the atomic selection. This provides a clear insight on the structural conformation of the protein throughout the simulation. Further RMSD analysis would also illustrate the equilibrated simulation if any, its fluctuations towards the end of simulation around thermal average structure. During simulation, changes of order from 1 to 3 Å for small, globular proteins are tolerable. The changes are much greater than the above specified value indicates large conformational change of the protein during simulation.
It is to be noted that if the simulation converges, the RMSD values stabilize around a fixed value. The increasing or decreasing pattern of RMSD of the protein at the end of the simulation, indexes that the system is not equilibrated. Ligand RMSD indicates the stability of the ligand with respect to the protein and its binding pocket. RMSD of protein-ligand complex is aligned on the protein backbone of the reference and later to which, the same for the ligand heavy atoms is measured. It is suggested that if the values are significantly larger than the RMSD of the protein, it is likely that the ligand has diffused away from its initial binding site [Citation18].
3.9 Molecular simulation analysis of AmEcR-20E complex
RMSD was drawn from the super-imposed conformational structure for AmEcR and 20E complex, it showed that AmEcR was stable in water between the distances of 2.3 and 2.6 Å variations (A). Analysis of AmEcR and 20E complex with potential deviation showed that it was simulated and had low potential energy ranging from −26100E to −26500E for the observed complex structure (B). Lowering in potential energy indicated for the increase in complexity in intermolecular bonding. The trajectories from 1 to 100 were super-imposed and checked for the movement of the complex structure. The structure alignment exhibited more complexity with respect to the water environment, since the environment was hydrophobic and the ligand had more effect in the internal bonding with AmEcR (C). The accurate prediction of protein-ligand binding free energies is a primary objective in computational drug design. Using the Optimise potential for liquid simulation (OPLS-2005) force field and obtain high correlation with experimental solvation free energies and low average unsigned errors for a majority of the functional groups [Citation19].
Fig. 6 Molecular dynamics of AmEcR-20E complex, A) RMSD graph B) Potential energy graph and C) 1–100 sample super-imposed structure of AmEcR and agonist 20E. AmEcR and 20E complex showed that AmEcR was stable in water between the distances of 2.3 and 2.6 Å variations. Potential deviation of this complex shows low potential energy ranging from −26100E to −26500E.

3.10 Molecular simulation analysis of AmEcR-Antagonist complex
Super imposed conformational structure for AmEcR and Etoxazole complex showed that AmEcR was stable in water between the distances of 1.8 and 2.3 Å (A) and ZINC20031812 showed from 1.9 to 2.3 Å variation (A). The mode of action of Etoxazole reveals the chitin biosynthesis inhibitor activity. The moulting defects were observed in fall armyworm Spodoptera frugiperda larvae [Citation20].
Fig. 7 Molecular simulation of AmEcR-Etoxazole complex, A) RMSD graph B) Potential energy graph and C) 1–100 sample super-imposed structure of AmEcR and antagonist Etoxazole. Super imposed conformational of AmEcR and Etoxazole complex shows the structural stability in water between the distances 1.8 and 2.3 Å and potential deviation ranging from −26000E to −26400E, lower potential energy confirms the binding stability.
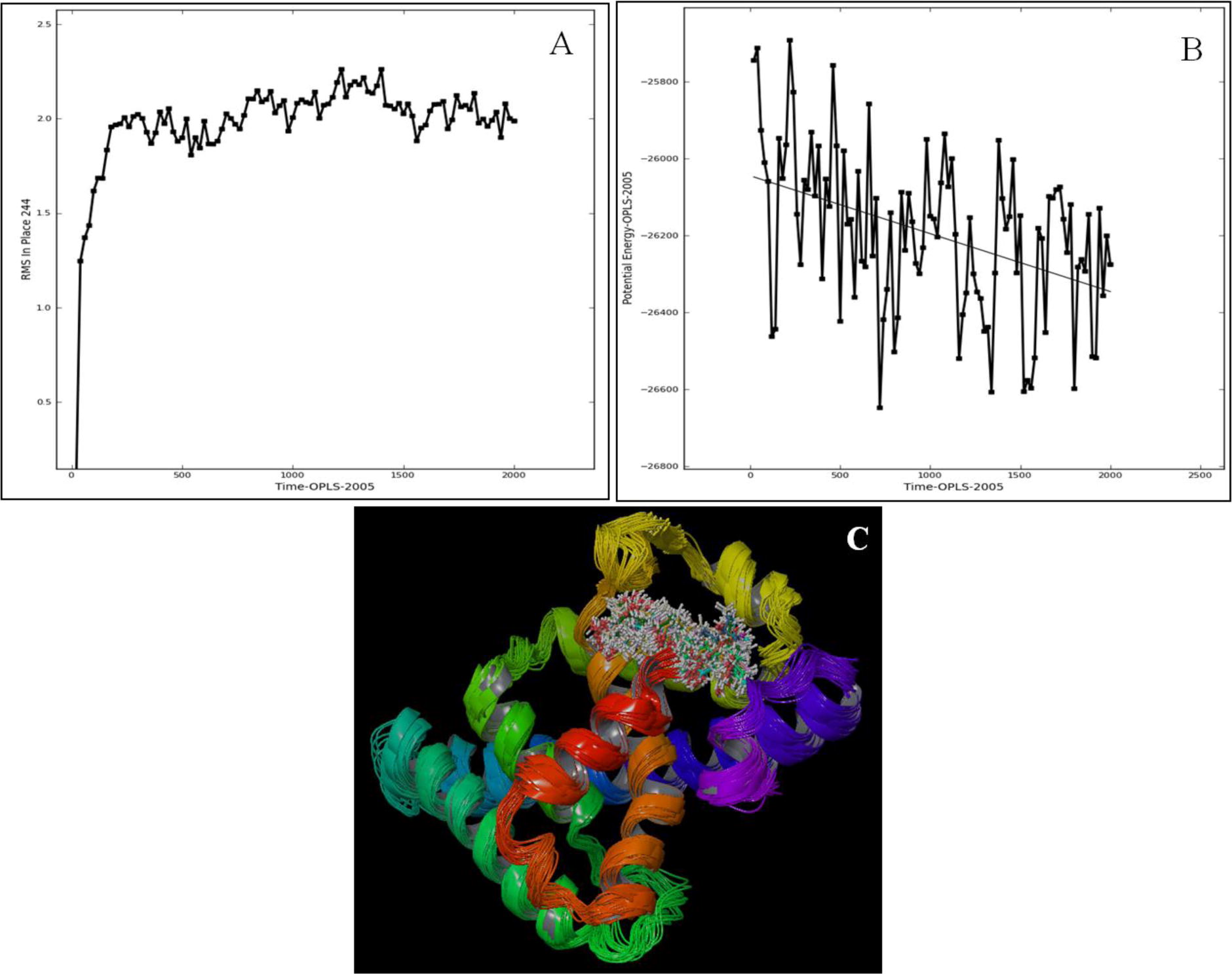
Fig. 8 Molecular simulation of AmEcR-ZINC20031812 complex A) RMSD graph B) Potential energy graph and C) 1–100 sample super-imposed structure of AmEcR and ZINC20031812. Super imposed structure for AmEcR and ZINC20031812 showed 1.9–2.3 Å variation which is stable in water and with lower potential energy ranging from −26000E to −26400E.
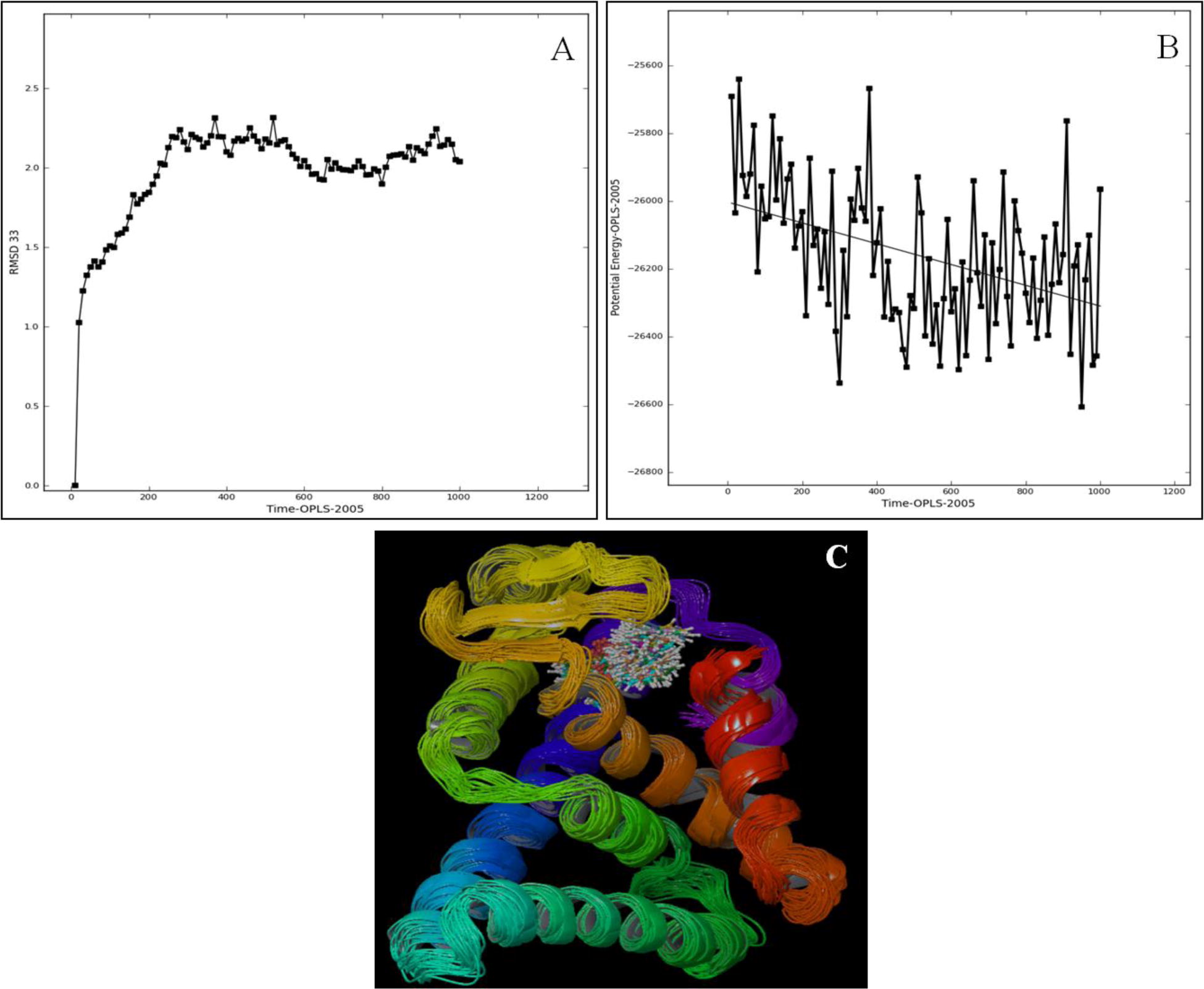
Simulated AmEcR and both antagonist complex with same potential deviation ranging from −26000E to −26400E, lower potential energy confirms the binding stability (B, B). The trajectories from 1 to 100 were super-imposed and checked for the movement of the complex structure. The structure alignment exhibited more complexity with respect to the water environment, since the environment was hydrophobic, and the ligand had more effect in the internal bonding with AmEcR (C, C). Chitin synthesis inhibitor activity of Etoxazole was tested against major insect pest of vegetables, beet armyworm (Spodoptera exigua), diamondback moth (Plutella xylostella), bean aphid (Aphis craccivora) and carmine spider mite (Tetranychus cinnabarinus) and its effective with LC50 also recorded [Citation21].
On comparing the RMSD value of agonist and antagonist indicates that they were stable in water with a range of distance from 2.3 to 2.6 Å, 1.8 to 2.3 Å and 1.9 to 2.3 Å with a variation over the time scale of 1 ps. Since Etoxazole and ZINC20031812 are antagonists, computationally they were more stable than 20E. In current work, we proposed a target selective potential compounds are capable for controlling specific insect pests, without causing considerable harm to other non-target organisms in the environment. In silico analysis enable more information on screening, possible mode of action and selecting a controlling agent before in vitro or in vivo studies. A novel caterpillar controlling agent named tebufenozide that poses very minimal hazard on non-target organisms and safe an ecosystem. It can be used on wide range of agricultural pests, and effectively replacing the environmentally toxic broad spectrum insecticides [Citation22]. Comparison of the EcR structures in complex with steroidal and non-steroidal ligands reveals radically different and only partially overlapping ligand-binding pockets that could not be predicted by molecular modeling and docking studies [Citation23]. The non-steroidal compound such as RH 5849 (1,2-dibenzoyl-1-tert-butylhydrazine) causes the premature initiation of moulting at all larval development stages of tobacco hornworm, Manduca sexta. The RH 5849 was 30 to >670 times as active as like the 20-hydroxyecdysone moulting hormone [Citation24]. By docking of 20E and dibenzoylhydrazines to the ecdysone receptor, a possible novel superposition of the natural and synthetic molecules are enabling for the designing of environment friendly insecticides [Citation15].
In the past, the control of arthropods depended mostly on chemical insecticides and these chemical pesticides are known to pollute the environment and also effects on non-target species. Chemical pesticides are not only depleting the nutritional value of our food, also it accumulates. Research has consistently found pesticide residues in food, by consuming it leads to myriad of diseases. The World Health Organization (WHO) estimates that there are 3 million cases of pesticide poisoning each year and up to 220,000 deaths, primarily in developing countries and children are more vulnerable to the chemical pesticides.
Now-a-days, there is more number of databases which provide thousands of plant based compound. These bioactive compounds may be potent in controlling the serious agricultural pest and also they are eco-friendly. These bioactive compounds may have allosteric regulation property, it can target the protein in other potential binding sites which may be effectively changes the conformational structure of protein and inhibits the binding of natural 20-hydroxyecdysone hormone in its dedicated pocket. The limonoids a compound belonging to tetranortriterpenoid group, in general, compounds belong to this group exhibits a wide range of biological activities like insecticidal, anti-feedant and growth regulating activity on insects [Citation25]. Cytotoxic effect of nimbolide (a limonoid) was analyzed on Sf9 (insect) cell lines, whereas the concentration at 9.8 μM acts fast on the Sf cells lines and induced disruption of plasma membranes [Citation26]. Also, Cucurbitacins B and D, isolated from seeds of Iberis umbellata (Cruciferae) have been shown to be responsible for the antagonistic activity in preventing the 20E induced morphological changes in Drosophila melanogaster BII permanent cell line [Citation27].
The larvae of many Lepidopteron species are major pests in agriculture. The major pest family belongs to Noctuidae, Pyralidae and Tortricidae. Ariadne merione belongs to the order Lepidoptera and family Nymphalidae. Although it is not considered as a serious pest, the in silico studies facilitates to maintain the data on target protein, compounds and an insight to design insect specific biopesticide.
4 Conclusion
AmEcR gene was amplified, sequenced and computational proteomics analysis were done to understand the mechanism of agonist and antagonist molecule in biocontrol perspective. Biocontrol method will be more precise due to the receptor specific antagonist designed using bioinformatic tools and this concept will be insect specific and it helps in preserving beneficial insect and the environment.
Acknowledgement
The authors are thankful to The Management, Karpagam University, Karpagam Academy of Higher Education, Coimbatore for providing all facilities required during this study.
References
- B.JayachandranM.HussainS.AsgariRegulation of Helicoverpa armigera ecdysone receptor by miR-14 and its potential link to baculovirus infectionJ Invertebr Pathol1142013151157
- D.Mané-PadrósF.Borràs-CastellsX.BellesD.MartínNuclear receptor HR4 plays an essential role in the ecdysteroid-triggered gene cascade in the development of the hemimetabolous insect Blattella germanicaMol Cell Endocrinol3482012322330
- C.S.ThummelFrom embryogenesis to metamorphosis: the regulation and function of Drosophila nuclear receptor superfamily membersCell831995871877
- W.W.ZhengD.T.YangJ.X.WangS.Q.SongL.I.GilbertX.F.ZhaoHsc70 binds to ultraspiracle resulting in the upregulation of 20-hydroxyecdsone-responsive genes in Helicoverpa armigeraMol Cell Endocrinol3152010282291
- H.MerzendorferL.ZimochChitin metabolism in insects: structure, function and regulation of chitin synthases and chitinasesJ Exp Biol2062003 4393-12
- G.SmaggheL.DecombelL.TirrySignificance of absorption, oxidation, and binding to toxicity of four ecdysone agonists in multi-resistant cotton leafwormArch Insect Biochem Physiol462001127129
- C.M.TiceSelecting the right compounds for screening: does Lipinski’s Rule of 5 for pharmaceuticals apply to agrochemicals?Pest Manage Sci572001316
- R.P.YadavK.S.IbrahimG.GurusubramanianN.SenthilkumarIn silico docking studies of non-azadirachtin limonoids against ecdysone receptor of Helicoverpa armigera (Hubner) (Lepidoptera: Noctuidae)Med Chem Res24201526212631
- J.J.IrwinB.K.ShoichetZINC – a free database of commercially available compounds for virtual screeningJ Chem Inf Model452005177182
- T.HaradaY.NakagawaM.AkamatsuH.MiyagawaEvaluation of hydrogen bonds of ecdysteroids in the ligand–receptor interactions using a protein modeling systemBioorg Med Chem17201158685873
- A.KasuyaY.SawadaY.TsukamotoK.TanakaT.ToyaM.YanagiBinding mode of ecdysone agonists to the receptor: comparative modeling and docking studiesJ Mol Model920035865
- T.IwemaA.ChaumotR.A.StuderM.R.RechaviI.M.L.BillasD.Moraset al.Structural and evolutionary innovation of the heterodimerization interface between USP and the ecdysone receptor EcR in insectsMol Biol Evol262009753768
- B.BordasI.BelaiA.LopataZ.SzantoInterpretation of scoring functions using 3D molecular fields. Mapping the diacyl-hydrazine-binding pocket of an insect ecdysone receptorJ Chem Inf Model472007176185
- Y.NakagawaV.C.HenrichArthropods nuclear receptors and their role in moultingFEBS J276200961286157
- J.M.WurtzB.GuillotJ.FagartD.MorasK.TietjenM.SchindlerA new model for 20-hydroxyecdysone and dibenzoylhydrazine binding: A homology modeling and docking approachProtein Sci9200010731084
- D.Tohidi-EsfahaniM.C.LawrenceL.D.GrahamG.N.HannanA.M.SimpsonR.J.HillIsoforms of the heteropteran Nezara viridula ecdysone receptor: protein characterisation, RH5992 insecticide binding and homology modellingPest Manage Sci67201114571467
- Bowers KJ, Chow E, Xu H, Dror RO, Eastwood MP, Gregersen BA et al. Scalable algorithms for molecular dynamics simulations on commodity clusters, Proceedings of the ACM/IEEE conference on supercomputing (SC06), Tampa, Florida; 2006. p. 11–17.
- Schrödinger release, Maestro-Desmond interoperability tools, version 3.6, Schrödinger. D.E. Shaw Research, New York; 2013.
- D.ShivakumarJ.WilliamsY.WuW.DammJ.ShelleyW.ShermanPrediction of absolute solvation free energies using molecular dynamics free energy perturbation and the OPLS force fieldJ Chem Theory Comput6201015091519
- R.NauenG.SmaggheMode of action of etoxazolePest Manage Sci622004379382
- Y.LiN.YangX.WeiY.LingX.YangQ.WangEvaluation of etoxazole against insects and acari in vegetables in ChinaJ Insect Sci142014114
- Carlson GR. Tebufenozide: a novel caterpillar control agent with unusually high target selectivity. Green chemical syntheses and processes. In: ACS symposium series, Pennsylvania; 2000. p. 8–17.
- I.M.L.BillasT.IwemaJ.M.GarnierA.MitschlerN.RochelD.MorasStructural adaptability in the ligand-binding pocket of the ecdysone hormone receptorLett Nat42620039196
- K.D.WingR.A.SlaweckiG.R.CarlsonRH-5849, nonsteroidal ecdysone agonist: effects on larval LepidopteraScience2411988470472
- A.RoyS.SarafLimonoids: overview of significant bioactive triterpenes distributed in plants kingdomBiol Pharm Bull292006191201
- E.CohenG.B.QuistadJ.E.CasidaCytotoxicity of nimbolide, epoxyazadiradione and other limonoids from neem insecticideLife Sci58199610751081
- L.DinanP.WhitingJ.P.GiraultR.LafontT.S.DhadiallaD.E.Cresset al.Cucurbitacins are insect steroid hormone antagonists acting at the ecdysteroid receptorBiochem J3271997643650