Abstract
Staphylococcal mastitis is a major and costly problem of dairy cattle all over the world. The objective of this study was to isolate and identify the main staphylococcal species causing bovine mastitis in 7 dairy herds located in 4 different areas of East and West Azerbaijan provinces, Iran. Of the 158 mastitic milk samples collected, 113 staphylococcal isolates were identified (71.5%) on the basis of cultural and biochemical features as well as by genus specific PCR. Then, species level identification of staphylococcal isolates was carried out using restriction fragment length polymorphism (RFLP) analysis of the gap gene (933 bp). On the basis of polymerase chain reaction-RFLP, 10 different patterns were identified. Of 113 isolates, 5 (4.4%) were Staphylococcus aureus and 108 (95.6%) were coagulase-negative staphylococci (CoNS). Overall, nine different species of CoNS were identified as: 44 Staphylococcus haemolyticus (40.7%), 17 Staphylococcus chromogenes (15.7%), 11 Staphylococcus epidermidis, Staphylococcus warneri and Staphylococcus cohnii each (10.2%), 6 Staphylococcus simulans (5.5%), 4 Staphylococcus hominis (3.7%), 3 Staphylococcus capitis (2.7%) and 1 Staphylococcus xylosus (0.9%). S. haemolyticus, S. chromogenes and S. warneri were the only species identified from clinical mastitis. No significant difference in staphylococcal IMI was found among the studied herds and regions. This study demonstrated that CoNS, especially S. haemolyticus and S. chromogenes, were predominant and thus be considered as emerging pathogens causing mastitis in the North West of Iran. Our results also revealed that the gap PCR-RFLP was useful for identifying staphylococcal isolates derived from bovine mastitis at species level.
1 Introduction
Staphylococci are the main etiological agents of mastitis in dairy cows [Citation1], comprises 45 species and 21 subspecies [Citation2]. Although Staphylococcus aureus has been described as one of the most important mastitis pathogens in cattle, coagulase-negative staphylococci (CoNS) are increasingly becoming recognized as etiologic agents associated with intramammary infections (IMI) in most countries [Citation3,Citation4]. However, their clinical/pathogenic relevance when cultured from milk remains a point of discussion. Some consider them as true mastitis pathogens with important virulence factors [Citation5], a high level of antimicrobial resistance [Citation6], and the ability to cause chronic infections [Citation7]. Others regard them as minor pathogens in dairy cows [Citation8,Citation9]. Today more than 15 CoNS species have been identified that cause IMI in dairy cows, but Staphylococcus chromogenes, Staphylococcus simulans, Staphylococcus xylosus, Staphylococcus epidermidis, Staphylococcus hyicus, and Staphylococcus haemolyticus are the most commonly isolated CoNS from bovine mastitis [Citation10,Citation11]. A number of other species have also been reported. Because of the increasing significance of CoNS in bovine mastitis, identification of CoNS at the species level is needed to develop effective control strategies for CoNS mastitis [Citation12]. Also, evaluating the epidemiology of individual CoNS species is of great importance to understand their respective significance [Citation13]. Currently the identification methods for staphylococci are under reassessment and molecular based methods are reported to be accurate alternatives and are superior to phenotypic identification of CoNS [Citation14,Citation15]. DNA sequence-base species identification is currently the most accurate method for CoNS identification and is considered as the gold standard [Citation16], but is expensive and time consuming. The polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) assay of gap gene, which encodes glyceraldehyde-3-phospahte dehydrogenase, has proved to be a simple, more reliable and reproducible method for differentiating staphylococcal species and allow the identification of 28 species within the genus [Citation17,Citation18]. Although the role of S. aureus as etiological agents of bovine mastitis has been elucidated previously [Citation19,Citation20], studies on the relative importance of specific CoNS in bovine mastitis have not been undertaken in Iran. Therefore, the overall aim of the present study was to improve the knowledge on prevalence and significance of different staphylococci species with emphasis on distribution of CoNS species.
2 Material and methods
2.1 Herds and cows
In total, 158 milk samples were collected from 31 clinical (recognized by physical examination and observable changes in the udder and the presence of abnormalities in the milk) and 127 subclinical (determined by CMT and bacteriological culture) mastitis of dairy cows during 2012. Seven herds (labeled A–G) located in four areas (including Jolfa, Khosrowshahr, Azarshahr, and Miandoab) of East and West Azerbaijan provinces from North West of Iran () were selected for this study: A (29 cows), B (24 cows), C (26 cows), D (23 cows), E (22 cows), F (12 cows), and G (22 cows). The farms were representative of Iranian dairy herds regarding cow number, milk production, and management routines (housing, feeding, and milking). In the year of the study, cows were in 3–4 parity, the mean herd size was 48 with a range from 25 to 70 Holstein cows, the average milk production was 27.8 kg/cow per day (range 25.2–30.4 kg/d). Cows on each herd were milked twice daily in a milking parlor. On all 7 farms, milkers did not wear gloves during the milking process. Post milking teat disinfection and dry cow therapy were practiced in all herds. Cows were housed in open shed with concrete floor and dried manure solids (DMS) were used as bedding for dairy cows.
2.2 Milk sampling and analysis
Milk samples were collected according to the procedures of the National Mastitis Council [Citation21]. Milk samples (15 ml) were taken into the sterile tube after cleaning of the teat ends using the alcohol soaked cotton and discarding the first streams of milk. Samples were shipped on ice to the laboratory, and bacteriological analysis was initiated within 24 h after sampling. 100 μl of milk was streaked on 5% sheep blood agar plates (Merck, Germany), and incubated aerobically at 37 °C for 18–24 h. Colonies suspected of being staphylococci were initially identified by their colony morphology and Gram staining. For Gram-positive cocci, catalase test was performed to distinguish catalase-negative Streptococcus spp. from catalase-positive Staphylococcus spp. Coagulase production by coagulase-positive staphylococci (CoPS) was examined using the tube coagulase method [Citation22]. Then, all initially identified isolates were further confirmed as Staphylococcus using genus specific oligonucleotide primers Sta I (5′-GGA ATA ACG TGA CAT ATT GTA-3′) and Sta II (5′-TTC ACT CGG TTT TGC TTG G-3′) previously described by Forsman et al. [Citation23]. DNA extraction from isolates was carried out by using the genomic DNA purification kit under the conditions described by the supplier (Fermentas, Germany). Then, isolates were stored in brain heart infusion broth with 15% glycerol at −70 °C until further identification.
2.3 PCR-RFLP of the gap gene
Species level identification of staphylococci isolates was carried out by PCR-RFLP of the gap gene. The primers GF-1 (5′-ATG GTT TTG GTA GAA TTG GTC GTT TA-3′) and GR-2 (5′-GAC ATT TCG TTA TCA TAC CAA GCT G-3′) were used to amplify the 933-bp gap gene fragment [Citation17]. The PCR was performed on Corbett thermal cycler (model CP2-003, Australia) in a final volume of 50 μl containing 6 μl of the DNA as the template, 1.25 U Taq DNA polymerase, 5 μl of 10× PCR buffer, 0.4 mM dNTPs, 0.8 μM of each primer, 3 mM MgCl2 and 17 μl distilled water. The PCR conditions were as follows: initial denaturation at 94 °C for 5 min, 40 cycles of amplification (DNA denaturation at 94 °C for 20 sec, annealing of primers at 55 °C for 30 sec, and extension at 72 °C for 40 sec), and final extension at 72 °C for 5 min. S. aureus ATCC 29213 was used as positive control. PCR products were analyzed by agarose gel electrophoresis in 0.5× Tris–borate–EDTA (TBE) buffer. The size of the amplicon was determined by comparison with the GeneRuler™ 100 bp plus DNA ladder (Fermentas, Germany).
The RFLP procedure was performed with 10 μl of the PCR products using AluI restriction endonuclease (Fermentas, Germany) according to the manufacturer’s recommendation. The resulting fragments were separated by electrophoresis on a 1% (wt/vol) agarose gel (sigma, Germany) at 80 V for 4 h and were visualized under UV light. The size of digested bands was determined using GeneRuler™ 50 bp DNA ladder (Fermentas, Germany). The banding patterns from the PCR-RFLP of gap gene were compared with known profiles from the Yugueros et al. [Citation17,Citation18], to determine the species of staphylococci isolates. To ensure the reproducibility of gap PCR-RFLP, banding patterns obtained from triplicate assays of all isolates was also compared.
2.4 Statistical analysis
Chi-square test was performed using SPSS Software version 22 (IBM SPSS Statistics for Windows, Armonk, NY, USA: IBM Corp.) to compare the staphylococcal IMI between the studied herds and regions. For the test, P < 0.05 was considered statistically significant.
3 Results
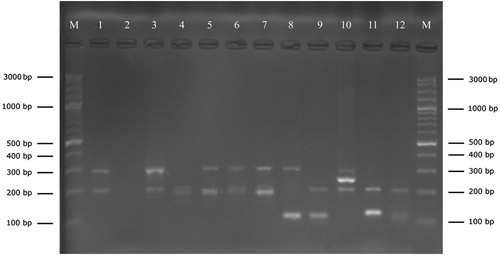
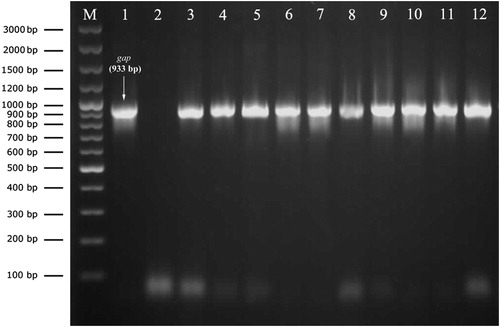
A total of 113 staphylococci were isolated from bovine mastitic milk samples, 20 (17.7%) of those from clinical and 93 (82.3%) from subclinical cases. Amplification with the genus specific PCR successfully confirmed the isolates as Staphylococcus spp. by amplification of DNA fragments ranging in size from 100 bp to 300 bp (). PCR amplification with the primer set of GF-1 and GR-2 produced a single amplicon of 933 bp in all staphylococci isolates ().
Among the 113 staphylococci milk isolates, 10 species were identified according to the restriction profile of the gap gene using AluI endonuclease (). The RFLP patterns were reproducible and generated the identical banding patterns by analyzing all isolates in triplicate. Overall, S. aureus (n = 5; 4.4%) and 9 different coagulase-negative staphylococci (CoNS) (n = 108; 95.6%) were identified. Among CoNS, S. haemolyticus (n = 44; 40.7%) and S. chromogenes (n = 17; 15.7%) were the most common findings followed by S. epidermidis, Staphylococcus warneri, and Staphylococcus cohnii (n = 11; 10.2% each), S. simulans (n = 6; 5.5%), S. hominis (n = 4; 3.7%), Staphylococcus capitis (n = 3; 2.7%) and S. xylosus (n = 1; 0.9%). The distribution of staphylococcal isolates within herds and detailed information about their incidence in clinical and subclinical cases is given in .
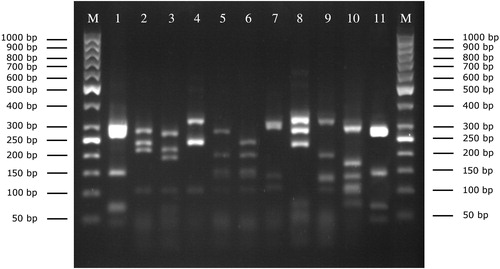
Table 1 Distribution of staphylococci species isolated from clinical and subclinical mastitis bovine milk samples on seven dairy herds (A–G).
Three species (S. haemolyticus, S. warneri, S. chromogenes) were found in both clinical and subclinical cases of mastitis, while seven species (S. aureus, S. cohnii, S. hominis, S. xylosus, S. epidermidis, S. capitis, and S. simulans) were found only in subclinical cases. S. haemolyticus, S. chromogenes, S. epidermidis and S. cohnii were the most common finding in subclinical mastitis.
Statistical analyses demonstrated that the staphylococcal IMI was not significantly different among the studied herds and regions.
4 Discussion
Staphylococcal mastitis are the most common and costly mammary disease of dairy cattle worldwide [Citation24]. In Iran, the prevalence of Staphylococcus spp. causing bovine mastitis has not been reported, so far. In this study, a total of 113 staphylococci isolates were identified in 158 milk samples (71.5%) by cultural as well as the genus specific PCR. This result is in agreement with the findings of Hashemi et al. [Citation25] and Atyabi et al. [Citation26] who introduced staphylococci as the most common isolated bacteria in other regions of Iran. Molecular assays targeting some housekeeping genes such as gap [Citation17,Citation18] have been used for reliably identifying and classifying staphylococci. In this study, gap PCR-RFLP assay was performed for species level identification of isolated staphylococci. According to the results, coagulase-negative staphylococci (CoNS) accounted for 68.35% while S. aureus accounted for 3.2% of intramammary infection (IMI). This indicates that CoNS play a prominent role in bovine mastitis. Similar finding on the growing importance of CoNS as mastitis pathogens have reported by others from different countries [Citation8,Citation27,Citation28]. However, the prevalence of CoNS mastitis varies between studies as reviewed by Pyorala and Taponen [Citation27]. The high percentage of CoNS IMI shows that current mastitis management in the studied herds (such as teat disinfection and dry cow therapy) is less successful in controlling CoNS IMI. There have been reports of a remarkably high frequency of CoNS infections in some herds (for unknown reasons), despite the use of preventive practices have substantially reduced the incidence of contagious major pathogens [Citation8,Citation29]. One speculation for the high presence of CoNS may be decreased teat sphincter patency as a consequence of high parity and late lactation which can result in CoNS being propelled into the mammary gland. Dried manure solids (DMS), as bedding in the studied herds, may also be a risk factor for CoNS mastitis. It has demonstrated that cows housed with organic or sand bedding had the most prevalence of CoNS IMI compared with cows housed with no bedding [Citation30].
Studies on CoNS from bovine milk show a wide variation with regard to species isolated most frequently. More recently, the predominant CoNS species isolated from bovine IMI are S. haemolyticus, S. chromogenes, S. epidermidis, S. simulans, and S. xylosus [Citation13,Citation31–Citation33] but other staphylococci commonly isolated include Staphylococcus sciuri, and S. cohnii [Citation9,Citation34]. In the current study, S. haemolyticus (40.7%) and S. chromogenes (15.7%) were the most frequently isolated CoNS, indicating that these are probably the predominant CoNS causing bovine mastitis in the North West of Iran. A possible explanation for this might be differences in pathogenic factors and genetic determinants of antibiotics which may cause change in the level of pathogenicity, sustainability and spreading of CoNS within and between animals. For example, haemolytic and proteolytic activity [Citation35], higher tolerance to post milking teat disinfection [Citation36,Citation37], and ability to evade the immune defense mechanisms [Citation38] might provide a possible colonization advantage for S. haemolyticus and/or S. chromogenes in mammary tissue. However, the present study was not carried out on diffusion of virulence and resistance genes and thus has no data on genetic variation among CoNS species. Therefore, the analysis of factors and comparison among CoNS is helpful in reduction of mastitis.
As results, S. simulans and S. xylosus was rarely detected in the current study. Compared to the results of the present study, previous studies, mostly conducted recently, have recovered higher percentages of S. simulans and/or S. xylosus [Citation9,Citation10,Citation13,Citation33,Citation34]. This might be due to the differences in the geographical distribution of CoNS, environmental and management conditions. According to Piessens et al. [Citation13], the origin of S. simulans and S. xylosus was primarily environmental. However, more research on this topic needs to be undertaken. In this study, S. cohnii was also found to be the commonly isolated CoNS species (10.2%). This result is in agreement with earlier study [Citation9] in which S. cohnii was isolated more frequently from bovine IMI, but its herd specific distribution as described in that study, was not observed in our study. S. warneri, S. hominis and S. capitis were together accounted for 16.7% of the CoNS isolates. Dairyman may be a potential source for colonization of these species in bovine udder tissue, as these species are commonly found living on the skin and the mucous membranes of humans [Citation39]. This hypothesis is supported by identification of S. epidermidis (a prominent member of the human skin flora) as the third most common CoNS (n = 11; 10.2%) responsible for IMI in this study. Thorberg et al. [Citation40] found S. epidermidis isolates with same pulsed-field gel electrophoresis (PFGE) types in samples from milk and milker’s skin, which indicate that S. epidermidis may be transmitted from milkers to cows.
In our study, we did not detect significant differences with respect to the occurrence of staphylococcal IMI among the studied herds as well as regions. This may be attributed to same management characteristics, housing environments, and also climate conditions. However, minor differences were seen in CoNS species distribution among the herds included in our study. The herd dependency of CoNS species distribution has been mentioned in other studies [Citation9,Citation10]. This indicates that unknown herd level factors or environmental conditions determine the establishment of particular staphylococcal species in a dairy herd. For example, it has demonstrated that post milking teat disinfection influence the distribution of CoNS species [Citation37]. Further studies on influence of herd-specific management programs and environmental factors on CoNS prevalence and distribution are required.
Thought most IMI caused by CoNS tend to remain subclinical and are left untreated, CoNS can also develop clinical mastitis and persistent intramammary infections [Citation7,Citation41]. In this study, S. haemolyticus, S. warneri and S. chromogenes were cultured from clinical bovine mastitis, underscoring the known pathogenicity of these species as mastitis pathogens in cattle. This is somewhat in line with the finding of Waller et al. [Citation33], where S. chromogenes, S. simulans and S. haemolyticus tended to be associated with clinical mastitis more often than other CoNS species.
In conclusion, the genotypic characterization of staphylococci isolates from bovine milk samples showed that coagulase-negative staphylococci (CoNS), especially S. haemolyticus and S. chromogenes, were a frequent cause of bovine intramammary infection. Thus, the study suggests the need for continuous monitoring of CoNS in herds, and special attention needs to be placed on the prevention and control of CoNS mastitis pathogens.
Acknowledgments
This work was supported by the Research Foundation of Urmia University, Urmia, Iran. The authors would like to thank Dr. B. Dalir-Naghadeh for his assistance in data analysis.
Notes
Peer review under responsibility of Faculty of Veterinary Medicine, Cairo University.
References
- N.UnalM.YildirimAntibiotic resistance profiles of staphylococci species isolated from milks, teat skins and noses mucous of cowsKafkas Univ Vet Fak Derg162010389396
- M.BergeronO.DauwalderM.GouyA.M.FreydiereM.BesH.MeugnierY.BenitoJ.EtienneG.LinaF.VandeneschS.BoissetSpecies identification of staphylococci by amplification and sequencing of the tuf gene compared to the gap gene and by matrix-assisted laser desorption ionization time-of-flight mass spectrometryEur J Clin Microbiol Infect Dis302011343354
- N.UnalS.AskarH.C.MacunF.SakaryaB.AltunM.YildirimPanton-Valentine leukocidin and some exotoxins of Staphylococcus aureus and antimicrobial susceptibility profiles of staphylococci isolated from milks of small ruminantsTrop Anim Health Prod442012573579
- S.TaponenS.PyoralaCoagulase-negative staphylococci as cause of bovine mastitis – not so different from Staphylococcus aureus?Vet Microbiol13420092936
- S.ZhangC.W.MaddoxCytotoxic activity of coagulase-negative staphylococci in bovine mastitisInfect Immun68200011021108
- P.J.Rajala-SchultzA.H.TorresF.J.DegravesW.A.GebreyesP.PatchaneeAntimicrobial resistance and genotypic characterization of coagulase-negative staphylococci over the dry periodVet Microbiol13420095564
- B.E.GillespieS.I.HeadrickS.BoonyayatraS.P.OliverPrevalence and persistence of coagulase-negative Staphylococcus species in three dairy research herdsVet Microbiol13420096572
- Y.H.SchukkenR.N.GonzalezL.L.TikofskyH.F.SchulteC.G.SantistebanF.L.WelcomeG.J.BennettM.J.ZurakowskiR.N.ZadoksCNS mastitis: nothing to worry about?Vet Microbiol1342009914
- K.SupreF.HaesebrouckR.N.ZadoksM.VaneechoutteS.PiepersS.De VliegherSome coagulase-negative Staphylococcus species affect udder health more than othersJ Dairy Sci94201123292340
- B.M.ThorbergM.L.Danielsson-ThamU.EmanuelsonK.Persson WallerBovine subclinical mastitis caused by different types of coagulase-negative staphylococciJ Dairy Sci92200949624970
- J.Y.ParkL.K.FoxK.S.SeoM.A.McGuireY.H.ParkF.R.RurangirwaW.M.SischoG.A.BohachDetection of classical and newly described staphylococcal superantigen genes in coagulase-negative staphylococci isolated from bovine intramammary infectionsVet Microbiol1472011149154
- A.A.SawantB.E.GillespieS.P.OliverAntimicrobial susceptibility of coagulase-negative Staphylococcus species isolated from bovine milkVet Microbiol13420097381
- V.PiessensE.Van CoillieB.VerbistK.SupreG.BraemA.Van NuffelL.De VuystM.HeyndrickxS.De VliegherDistribution of coagulase-negative Staphylococcus species from milk and environment of dairy cows differs between herdsJ Dairy Sci94201129332944
- A.CapurroK.ArturssonK.P.WallerB.BengtssonH.Ericsson-UnnerstadA.AspanComparison of a commercialized phenotyping system, antimicrobial susceptibility testing, and tuf gene sequence-based genotyping for species-level identification of coagulase-negative staphylococci isolated from cases of bovine mastitisVet Microbiol1342009327333
- O.JoussonD.Di BelloM.VanniG.CardiniG.SoldaniC.PrettiL.IntorreGenotypic versus phenotypic identification of staphylococcal species of canine origin with special reference to Staphylococcus schleiferi subsp. coagulansVet Microbiol1232007238244
- R.N.ZadoksJ.L.WattsSpecies identification of coagulase-negative staphylococci: genotyping is superior to phenotypingVet Microbiol13420092028
- J.YuguerosA.TempranoB.BerzalM.SanchezC.HernanzJ.M.LuengoG.NaharroGlyceraldehyde-3-phosphate dehydrogenase-encoding gene as a useful taxonomic tool for Staphylococcus sppJ Clin Microbiol38200043514355
- J.YuguerosA.TempranoM.SanchezJ.M.LuengoG.NaharroIdentification of Staphylococcus spp. by PCR-restriction fragment length polymorphism of gap geneJ Clin Microbiol39200136933695
- H.D.SaeiM.AhmadiK.MardaniR.A.BatavaniMolecular typing of Staphylococcus aureus isolated from bovine mastitis based on polymorphism of the coagulase gene in the north west of IranVet Microbiol1372009202206
- H.MomtazE.TajbakhshE.RahimiM.MomeniCoagulase gene polymorphism of Staphylococcus aureus isolated from clinical and sub-clinical bovine mastitis in Isfahan and Chaharmahal va Bakhtiari provinces of IranComp Clin Path202011519522
- National Mastitis Council (NMC). Laboratory handbook on bovine mastitis. Madison, Wisconsin: Natl Mastitis Counc Inc; 1999.
- Quinn PJ, Carter ME, Markey BK, Carter GR. Clinical Veterinary Microbiology. 2nd ed. UK, London: Mosby publish ring; 1998.
- P.ForsmanA.Tilsala-TimisjarviT.AlatossavaIdentification of staphylococcal and streptococcal causes of bovine mastitis using 16S–23S rRNA spacer regionsMicrobiology143Pt 11199734913500
- G.LeitnerO.KrifucksM.D.KiranN.BalabanVaccine development for the prevention of staphylococcal mastitis in dairy cowsVet Immunol Immunopathol14220112535
- M.HashemiM.KafiM.SafdarianThe prevalence of clinical and subclinical mastitis in dairy cows in the central region of Fars province, south of IranIran J Vet Res122011236241
- N.AtyabiM.VodjganiF.GharagozlooA.BahonarPrevalence of bacterial mastitis in cattle from the farms around TehranIran J Vet Res720067679
- S.PyoralaS.TaponenCoagulase-negative staphylococci-emerging mastitis pathogensVet Microbiol134200938
- J.K.El-JakeeN.E.ArefA.GomaaM.D.El-HaririH.M.GalalS.A.OmarA.SamirEmerging of coagulase negative staphylococci as a cause of mastitis in dairy animals: an environmental hazardInt J Vet Sci Med120137478
- S.PiepersL.De MeulemeesterA.de KruifG.OpsomerH.W.BarkemaS.De VliegherPrevalence and distribution of mastitis pathogens in subclinically infected dairy cows in Flanders, BelgiumJ Dairy Res742007478483
- J.D.FergusonG.AzzaroM.GambinaG.LicitraPrevalence of mastitis pathogens in Ragusa, Sicily, from 2000 to 2006J Dairy Sci90200757985813
- Y.D.TremblayD.LamarcheP.CheverD.HaineS.MessierM.JacquesCharacterization of the ability of coagulase-negative staphylococci isolated from the milk of Canadian farms to form biofilmsJ Dairy Sci962013234246
- P.AjitkumarH.W.BarkemaR.N.ZadoksD.W.MorckF.J.van der MeerJ.De BuckHigh-resolution melt analysis for species identification of coagulase-negative staphylococci derived from bovine milkDiagn Microbiol Infect Dis752013227234
- K.P.WallerA.AspanA.NymanY.PerssonU.G.AnderssonCNS species and antimicrobial resistance in clinical and subclinical bovine mastitisVet Microbiol1522011112116
- Y.FreyJ.P.RodriguezA.ThomannS.SchwendenerV.PerretenGenetic characterization of antimicrobial resistance in coagulase-negative staphylococci from bovine mastitis milkJ Dairy Sci96201322472257
- M.BochniarzW.WawronHaemolytic and proteolytic activity of coagulase-negative staphylococci isolated from mastitis cowsPol J Vet Sci1520126165
- V.PiessensS.De VliegherB.VerbistG.BraemA.Van NuffelL.De VuystM.HeyndrickxE.Van CoillieCharacterization of coagulase-negative staphylococcus species from cows’ milk and environment based on bap, icaA, and mecA genes and phenotypic susceptibility to antimicrobials and teat dipsJ Dairy Sci95201270277038
- T.QuirkL.K.FoxD.D.HancockJ.CapperJ.WenzJ.ParkIntramammary infections and teat canal colonization with coagulase-negative staphylococci after post milking teat disinfection: species-specific responsesJ Dairy Sci95201219061912
- M.ChafferG.LeitnerM.WinklerA.GlickmanO.KrifucksE.EzraA.SaranCoagulase-negative staphylococci and mammary gland infections in cowsZentralbl Veterinarmed B461999707712
- N.NagaseA.SasakiK.YamashitaA.ShimizuY.WakitaS.KitaiJ.KawanoIsolation and species distribution of staphylococci from animal and human skinJ Vet Med Sci642002245250
- B.M.ThorbergI.KuhnF.M.AarestrupB.BrandstromP.JonssonM.L.Danielsson-ThamPheno- and genotyping of Staphylococcus epidermidis isolated from bovine milk and human skinVet Microbiol1152006163172
- S.TaponenJ.KoortJ.BjorkrothH.SaloniemiS.PyoralaBovine intramammary infections caused by coagulase-negative staphylococci may persist throughout lactation according to amplified fragment length polymorphism-based analysisJ Dairy Sci90200733013307