Abstract
In Egypt, two distinct genetic groups of HPAI H5N1 viruses are co-circulating: classic 2.2.1/C sub-clade and antigenic drift variant 2.2.1.1 clade isolated from vaccinated poultry flocks. The response of chicken innate immunity to both genotypes is not investigated, so far. In this study, expression of immune related genes (IL1b, IL4, IL6, IL8, IL10, IL18, IFNα and IFNγ) after infecting chicken macrophage cell line (HD11) and chicken peripheral blood Mononuclear cells (PBMC) with a classic and a variant strains was assayed using quantitative reverse-transcription real-time polymerase chain reaction assays (qRT-PCR). In HD11, the variant strain induced higher levels of IL1b and IL8 at 6 hours post infection (hpi), IL4 at 24 / 48 hpi and IFNα at 48 hpi than the classic strain. Conversely, the classic strain induced about 10-fold increase of IFNγ at 24 and 48 hpi and the virus replicated at higher level than the variant strain. The results of PBMC infection were similar to that reported from HD11 except for IFNγ gene expression that was higher at variant strain infected cells than that infected with the classic strain. After 24hpi skewing the innate immune response toward anti-inflammatory (humoral-associated) cytokines was different between HD11 (through IL4) and PBMC (through IL10). To sum up, the classic strain produced less cytokines which may indicate adaptation to evade the recognition by the innate immune system and explain its higher pathogenicity.
1 Introduction
Highly pathogenic avian influenza virus subtype H5N1 (HPAIV H5N1) is a worldwide devastating disease of poultry, which presents a potential pandemic threat [Citation1]. Since its emergence in 1997 in Hong Kong, the virus spread to more than 60 countries and finally became endemic in poultry populations at Bangladesh, China, Egypt, Indonesia and Viet Nam [Citation2]. In case of Egypt, losses in poultry industry since 2006 was estimated to exceed $1 billion due to culling or death of over 30 million birds [Citation3]. According to the WHO, the virus was spilled over to 173 persons and caused deaths in 63 patients by the 26th of November, 2013 [Citation4]. Vaccination of commercial poultry against the HPAIV H5N1 using different H5 vaccines was a milestone in the control of the disease in Egypt. The reduced number of outbreaks after the first wave in 2006 in poultry was attributed to the effectiveness of these vaccines to interrupt the circulation of the virus particularly in the commercial sector [Citation3]. Since 2007, a dramatic increase in the number of infected flocks despite vaccination was reported [Citation5].
Phylogenetic analyses of the Egyptian H5N1 viruses indicated co-circulation of two distinct genetic groups. The first group belongs to the 2.2.1/C subclade, also known as classic group, is very close to the predecessor 2.2.1 viruses introduced into Egypt in early 2006. These viruses were isolated from non-vaccinated backyard birds as well as from human [Citation6]. They were also able to induce clinical disease and mortality in improperly vaccinated chickens in small-scale commercial farms [Citation7]. The second genetic group classified separately in a unique 2.2.1.1 clade. Viruses in this clade represent the antigenic drift variants isolated from vaccinated birds and harbour major changes in immunogenic epitopes of the hemagglutinin (HA) protein [Citation6]. Experimental challenge studies showed that the classic group caused mild clinical signs in chickens vaccinated with homologous or heterologous H5 vaccines; however it was excreted for long periods of time. On the contrary, the variant strains caused up to 100% mortality in chickens vaccinated with heterologous vaccines and virus excretion was limited in birds vaccinated with homologous H5N1 vaccines [Citation8]. Accordingly, since 2011 no variant virus was isolated and the classic strains are the predominant genotype in Egypt due to probably adoption of more genetically related homologous vaccines in the commercial poultry [Citation6].
Much emphasis has been placed on the humoral immunity but the response of innate immune system of chickens to infections with H5N1 strains has not been adequately studied [Citation9]. The current dogma of the immunology states that the innate immune response is the first line of defence of a host against microbial invasion [Citation10]. An essential component of the innate immune system is cytokines which are triggered upon stimulation of host-cells with a micro-organism. They orchestrate innate and adaptive antiviral defence mechanisms to eliminate (e.g. influenza virus) infections from the host [Citation11]. According to their function, three classes of cytokines are mostly important (1) proinflammatory cytokines such as interleukin-1β (IL-1β), Interleukin-6 (IL6), IL8 and tumour necrosis factor-α (TNF-α) that play a role in the induction of inflammation during the course of infection, (2) T-Helper 1 (Th1) associated cytokines including IL18 and IFNγ that regulate and induce cell mediated immune (CMI) response and (3) anti-inflammatory/Th2 cytokines like IL4 and IL10 that involved in the induction and regulation of humoral immune response [Citation9]. In comparison to mammals, repertoire of cytokines in chicken was not fully understood until recently. A considerable number of chicken immune-related molecule orthologs have been identified and quantification of cytokine messenger RNA (mRNA) expression levels using quantitative real-time reverse-transcription polymerase chain reaction (qRT-PCR) improved our knowledge on the host-virus interaction [Citation12]. In human or mammal models, extensive literatures have been published on the molecular viral mechanisms involved in the H5N1 pathogenesis [Citation13] but very little is known about the host-influenza-interaction in chickens particularly the regulation of the innate immune response by HPAIV H5N1 immune-escape variants.
In the present study, HD11cell line and chicken PBMCs were used to study the regulation of cytokines upon infections with a classic and a variant HPAI strains isolated from chickens in Egypt.
2 Materials and methods
2.1 H5N1 strains propagation
Two viruses were obtained from the influenza virus repository of the Reference Laboratory for Quality Control on Poultry Production (RLQP), Egypt. A/chicken/Egypt/0963S-NLQP/2009(H5N1), GenBank accession number http://ncbi-n:HQ198269 belongs to the variant 2.2.1.1 clade and A/chicken/ Egypt/121/2012(H5N1), GenBank accession number http://ncbi-n:JQ858483 belongs to the classic 2.2.1/C subclade. Both viruses were propagated in 9 day-old specific pathogen free embryonated chicken eggs according to the standard protocol [Citation14]. All procedures were performed in BSL3 laboratory facilities at the National Institute of Animal Health (NIAH), Japan. Viral titters were expressed as mean tissue culture infectious dose (TCID50) using HD11 cell line for each strain according to Reed and Muench [Citation15].
2.2 HD11 cell line propagation and infection
HD11 cells were kindly provided by Dr. John Adams (the Cedars-Sinai Medical Centre, Los Angeles, CA, USA). Cell were counted and diluted in 2 mL growth media per well of 6-well plates to get 1.5×106 cell per well and incubated for 24 hrs to form a confluent sheet. After 24 hrs the media were removed and cells were washed with PBS. Cells were collected from one well of 6 well plate with trypsin and were counted to calculate the dose of the infection. A total volume of 0.5mL infection media contain the filtered viral allantoic fluid (0.05TCI50/cell) were added to every well and incubated for 1 hour with gently shaking every 15 min then the media was removed and cells were washed with PBS followed by addition of 2 mL of MEM medium (Gibco; Carlsbad, CA, USA). Plates were incubated at 37 °C in 5% CO2 incubator. HD11 cultures were used in parallel without the addition of viral allantoic fluid (VAF) as negative control. Samples (cells and supernatant) were collected at 6, 24 and 48 hr post infection (hpi) for RNA extraction as described below.
2.3 PBMC isolation, culturing and infection
Heparinized blood was collected form 10 week-old chickens and mixed with equal volume of PBS, then layered drop by drop over equal volume of Ficoll-Paque (GE health care, Sweden) followed by centrifugation at 400×g for 30 min without brake. PBMC were removed at the mononuclear cell interface carefully and washed twice with PBS and then subjected to centrifugation at 100×g for 10 min. The resultant pellet was re-suspended in complete RPMI media (Gibco, Carlsbad, CA, USA) containing 10% FBS. PBMC were grown overnight at cell density 107 per well of 12-well tissue culture plates and incubated at 37 °C in 5% CO2 incubator. After overnight growth, non-adherent cells were removed by washing the monolayers with PBS to enrich the cultures for adherent macrophages, monocytes and dendritic cells. One PBMC culture was trypsinized and the cells were counted to calculate the required amount of VAF to get infection dose of 0.05TCID50/cell. Growth media was replaced with infection media containing complete RPMI but without serum and with the addition of filtered VAF. Negative control PBMC cultures without the addition of VAF were incubated in parallel. Culture plates were gently shaken every 15 min for 1 hr then the media was replaced with RPMI supplemented with 0.2% FBS. Cultures were incubated at 5% CO2 and RNA was extracted from the cell monolayer and supernatant at 6, 24, and 48hpi.
2.4 Total RNA extraction
Total RNA was isolated from infected and control HD11 and PBMC cells and supernatant at each time point using RNeasy mini RNA Purification kit and DNase treatment with QIAGEN®™ RNA purification kits to purify RNA from DNA contamination following manufacturer’s instructions (Qiagen Inc., Valencia, CA, USA). RNA in each sample was quantified Using NanoDrop-1000 (Thermoscientific, Wilmington, DE).
2.5 qRT-PCR
qRT-PCR was performed using Quantitect probe RT-PCR (Qiagen, Inc. Valencia, CA, USA) according to the manufacturer recommendations. Primers and probes were selected for amplification of IL1b, IL6 and IFNγ [Citation16], IL4 and IL10 [Citation17], IL8 and IL18 [Citation18], and IFNα [Citation19] as shown in . qRT-PCR runs were performed using 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). PCR conditions were the same for each targeted gene of different cytokines as follows: 30 min at 50 °C, 95 °C for 15min, followed by 40 cycles of 95 °C for 15 s, 60 °C for 1min. For detection of AIV H5 [Citation20], the thermoprofile was 30 min at 50 °C, 95 °C for 15 min, followed by 40 cycles of 95 °C for 10 s, 54 °C for 30 s and 72 °C for 30 sec.
Table 1 Sequences of the primers and probes used in quantitative qRT-PCR.
Absolute quantification for viral RNA was calculated using tenfold-serially diluted RNA from known TCID50 classic and variant strains. The standard curves were created automatically by plotting the threshold level of fluorescence (Ct values) against the TCID50 for every dilution. The Ct values of the unknown samples were then compared to the Ct values of the standards. The starting template quantities for the unknown samples could be estimated in the present study using standard curve generated with every PCR run. For relative quantification of cytokines mRNA, amplification data of cytokines were normalized against 28s RNA [Citation16] and fold change of cytokines mRNA gene expression of infected cells compared to non-treated cells was calculated as previously published [Citation21].
3 Results
3.1 Virus titration
Serial titrations of both viral strains were performed in HD11 cell line and TCID50 titres were determined. The titer of the variant strain was calculated as 4×106.4 TCID50/mL while the classic strain was 4×105.6 TCID50/mL. The variant strain grew at lower levels than the classic strain in HD11 at all-time points reaching 105.5 and 106.8 TCID50/mL at 48 hpi for the variant and classic strains, respectively (data not shown). While in PBMCs, both viruses grew at similar TCID50 levels at different time point (data not shown).
3.2 Cytokines mRNA gene expression using HD11 cell line ()
Table 2 Cytokines mRNA fold change at 6, 24 and 48 h after infection of HD11 cell line with 0.05 TCID50 per cell.
Cytokines mRNA gene response was detected at 6, 24 and 48 hpi after infection using qRT-PCR (). The pro-inflammatory cytokines IL1b and IL8 were expressed at high levels and early at 6 hpi, while expression of IL4 and IFNγ increased later at 24 and 48 hpi compared with the non-infected cells. At 6 hpi, the variant strain increased the IL8, IL1b, IL4 and IL10 expression levels by 6.5, 4.2, 1.1 and 1.0-fold, respectively compared to those levels induced by the classic strain. Likewise, higher folds of about 1.0 and 7.1 at 24 hpi and 3.9 and 5.7 at 48 hpi for IL1b and IL4, respectively were obtained by the variant strains. Conversely, at 24 and 48 hpi about 18.0 and 14.0 higher folds of IFNγ respectively were induced by the classic strains than those obtained by the variant strain. IFNα gene expression levels were increased only at 48 hpi by the variant strain infected cells while other cytokines showed no marked changes ().
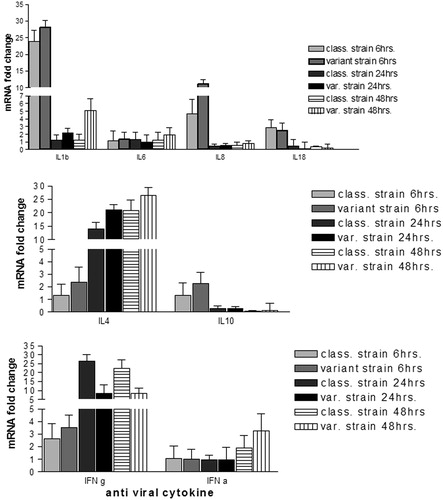
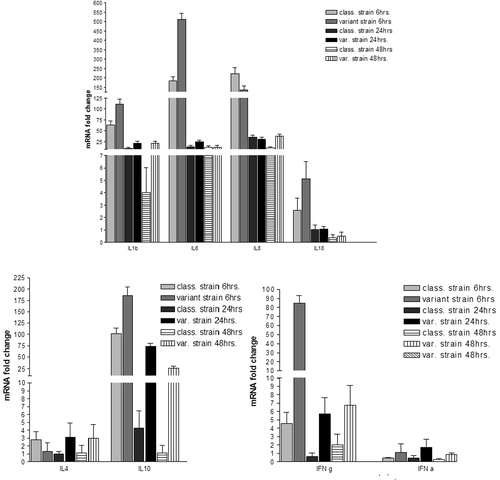
3.3 Cytokines mRNA gene expression using PBMC cell culture ()
Table 3 Cytokines mRNA fold change at 6, 24 and 48 h after infection of PBMC with 0.05 TCID50 per cell.
The level of cytokines mRNA gene expression markedly upregulated by both strains at each time point particularly IL1b, IL6 and IL8 at 6 hpi (). The variant strain at 6hpi elicited 48.1 to 322.4 folds higher of the IL1b, IL6, IL10 and IFNγ than the classic strain. Conversely, at 6 hpi the classic strain elicited 1.5 and 82.9 folds of IL4 and IL8, respectively higher than the variant strain. At 24hpi and 48hpi, higher levels of expression of most of cytokines induced by the variant strain than those induced by the classic strain were observed (). IL8 showed higher mRNA gene expression after infection of PBMC with the classical strain than that infected with variant strain at 6 hpi but at 48 hpi the variant strain infected PBMC showed higher IL8 mRNA gene expression (). PBMC infected with the variant strain showed marked higher IFNγ mRNA gene expression than that infected with the classic strain. The classic strain did not regulate IFNα mRNA gene expression similar to the negative control cells but infected PBMC with the variant strain was slightly upregulated ().
4 Discussion
Virulence of HPAIV H5N1 can be modulated by the virus-host immune system interaction [Citation22,Citation23]. Innate immune responses via interferon and other cytokines can limit or eliminate influenza virus infections. Variations of cytokines expression’ pattern between influenza virus serotypes (e.g. H5N1 and H1N1) [Citation24] or even within the same subtype (e.g. H5N1 of wild bird vs. of chickens origins) were intensively elucidated in several literatures [Citation12]. Macrophages are an important cellular component of the immune system and play a primary role in the development of both innate and adaptive immune responses. The use of chicken macrophage cells is an optimal system to study the interaction between HPAIV H5N1 and the immune system of birds, nevertheless only limited numbers of studies have been published [Citation25].
In this study, in HD11 cell line both virus genotypes up-regulated the expression level of cytokines, particularly at 6 hpi, compared to the negative control cells. At early stage of infection (6hpi) expression levels of the pro-inflammatory cytokines IL1b and IL8 were remarkably increased, but at later stages of infection (24 and 48hpi) the IL4 and IFNγ levels were the highest, while IFNα expression showed no marked change at early stage. Such results coincide with that of Watanabe et al. [Citation26] who recorded up-regulation of IL1b, IL8, IFNγ and IL18 in HD11 cell line infected with an HPAIV H5N1 [Citation27]. Also, low expression of IFNα was correlated with high viral titre and prolonged shedding time in chickens [Citation28]. Different expression patterns between both viruses were observed. Generally, the variant strain elicited higher IL1b, IL4, IL8 and IFNα but the classic strain induced higher IFNγ. This may explain the low replication titter of the variant strain in HD11and the positive correlation between viral titre and IFNγ expression recorded in vitro [Citation29]. These data support the field observation of the higher virulence of classical strain comparing to variant strain. As the variant strain induced higher innate immune response that represented by higher IL1b,IL8 and IFNα and the immunity skewed toward humeral immune response represented by IL4 that can efficiently reduce the pathogenicity of the virus and its titre, while classical strain induced lower innate immunity with higher IFNγ that correlated with higher viral titre. These finding agree with that of Friesenhagen et al. [Citation32], who suggested that much stronger inflammatory response of human macrophages to infection with low pathogenic virus than highly pathogenic avian influenza.
Avian species have a similar but not identical network of macrophages and dendritic cells (DC) to mammalian counterpart that was involved in uptake of foreign antigens [Citation29]. In PBMC, results of cytokines mRNA gene expression revealed that most of tested cytokines markedly elevated early at 6 hpi. That is expected because dendritic cells compared to other antigen presenting cells (macrophage and B cells) can effectively capture and process antigens, express higher level of MHC and co-stimulatory molecules on their surface and also activate naïve T cells [Citation30]. The results of PBMC infection almost equal to that reported from HD11 except for IFNγ gene expression that is higher at variant strain infected cells than that infected with classic strain; nonetheless both viruses grew at a similar level in PBMC. In mammals, the IL4 and IL10 play an important role in the promotion of Th2 responses (humoral immune response) and inhibition of the proinflammatory and Th1 response [Citation31]. This study indicated that after 24hpi skewing toward humoral immune response is essential and differs in HD11 (through IL4) and PBMC (through IL10) that was better marked with variant strain comparing to classical strain. Different expression levels of cytokines between HD11 were previously observed using two different pathotypes of H5N1 [Citation26]. It is worth mentioning that immune response to HPAIV H5N1 varies according the gene constellation of the virus [Citation26] and both Egyptian genotypes have specific genetic markers in all gene segments including the NS1 [Citation6] which is known to be the major viral host-immune system regulator. Thus, the role of internal proteins on different patterns of cytokine expression elicited by both strains in this study should be further elucidated. Generally, innate immune response differs after infection with low pathogenic AIV (LPAIV) and HPAIV [Citation26]. Friesenhagen et al. [Citation32] found that LPAIV induced stronger inflammatory responses in human macrophages than HPAIV which may facilitate wide spread and systemic progression of the later. From various field observations (non published data) variant strain induced lower pathogenicity and delayed onset of death in infected chicken comparing to classical strain infection. Here we investigate the difference in host immune response at the level of immune cells to both strains. Our data revealed that higher innate immune response (IL1b,IL6,IL8 and IFNα) with variant strain infections compared to classical strain infection. These results explain the lower pathogenicity recorded for variant strain may be due to efficient higher innate immune response that in turn skewed to higher humeral immune response (IL4 and IL10). Such mechanism can reduce the variant strain pathogenicity compared to classical strain infection.
5 Conclusion
In conclusion, the immune-escape HPAIV H5N1 elicited higher innate immune response than the “classic” virus. We propose that the capability of the variant virus to induced severe morbidity and mortality among poultry populations across Egypt is probably due to its ability to escape immune response elucidated by vaccination due to antigenic variation. Such variation is thought to affect its epitopes while triggering of higher innate immune response which do not play a significant role in this case. The classic strain produced less cytokine (in magnitudes and types) and it was able to replicate to high titter in HD11 and PMBC, which may indicate adaptation to evade the recognition by the innate immune system in case of infection. Ultimately, there will be two queries in a real need to be investigated; the in-vivo expression of cytokines and the immunogenicity / pathogenicity of the variant strain compared to the classic strain.
Acknowledgements
The authors are grateful to Dr. Tsunkuni from the Influenza and Prion Disease Research Centre Team, National Institute of Animal Health, Japan for their excellent technical assistance. The authors are also thankful to RLQP team. Special thanks are to Dr. Abdelwhab E.M., Institute of Molecular Biology, the Federal Research Institute for Animal Health / Germany for his valuable comments on the manuscript.
Notes
Peer review under responsibility of Faculty of Veterinary Medicine, Cairo University.
References
- J.S.PeirisM.D.de JongY.GuanAvian influenza virus (H5N1): a threat to human healthClin Microbiol Rev202007243267
- FAO. Approaches to controlling, preventing and eliminating H5N1 highly pathogenic avian influenza in endemic countries. Animal Production and Health Paper; No. 171. Rome; 2011.
- E.M.AbdelwhabH.M.HafezAn overview of the epidemic of highly pathogenic H5N1 avian influenza virus in Egypt: epidemiology and control challengesEpidemiol Infect1392011647657
- WHO. Situation updates – Avian influenza. Available online at: <http://www.who.int/influenza/human_animal_interface/avian_influenza/archive/en/>; 2013; (Accessed May 20, 2013).
- E.F.El-ZoghbyM.M.AlyS.A.NasefM.K.HassanA.S.ArafaA.A.SelimS.G.KholousyW.H.KilanyM.SafwatE.M.AbdelwhabH.M.HafezSurveillance on A/H5N1 virus in domestic poultry and wild birds in EgyptVirol J102013203
- E.M.AbdelwhabA.S.ArafaJ.StechC.GrundO.StechM.Graeber-GerberdingM.BeerM.K.HassanM.M.AlyT.C.HarderH.M.HafezDiversifying evolution of highly pathogenic H5N1 avian influenza virus in Egypt from 2006 to 2011Virus Genes4520121423
- E.F.El-ZoghbyA.S.ArafaW.H.KilanyM.M.AlyE.M.AbdelwhabH.M.HafezIsolation of avian influenza H5N1 virus from vaccinated commercial layer flock in EgyptVirol J92012294
- E.M.AbdelwhabC.GrundM.M.AlyM.BeerT.C.HarderH.M.HafezMultiple dose vaccination with heterologous H5N2 vaccine: immune response and protection against variant clade 2.2.1 highly pathogenic avian influenza H5N1 in broiler breeder chickensVaccine29201162196225
- Kaiser P, Stäheli P. Avian cytokines and chemokines. In: Davison F, Kaspers B, Schat KA (Eds.), Avian immunology. 1st ed., London, UK: Academic Press; 2008. p. 203–22.
- O.TakeuchiS.AkiraInnate immunity to virus infectionImmunol Rev22720097586
- P.KaiserT.Y.PohL.RothwellS.AveryS.BaluU.S.PathaniaS.HughesM.GoodchildS.MorrellM.WatsonN.BumsteadJ.KaufmanJ.R.YoungA genomic analysis of chicken cytokines and chemokinesJ Interferon Cytokine Res252005467484
- L.SarmentoC.L.AfonsoC.EstevezJ.WasilenkoM.Pantin-JackwoodDifferential host gene expression in cells infected with highly pathogenic H5N1 avian influenza virusesVet Immunol Immunopathol1252008291302
- W.C.YuR.W.ChanJ.WangE.A.TravantyJ.M.NichollsJ.S.PeirisR.J.MasonM.C.ChanViral replication and innate host responses in primary human alveolar epithelial cells and alveolar macrophages infected with influenza H5N1 and H1N1 virusesJ Virol85201168446855
- OIE. Avian influenza. Available online at: <http://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/2.03.04_AI.pdf>; 2009.
- L.G.ReedH.MuenchA simple method of estimating fifty percent endpointsAm J Hyg271938493497
- P.KaiserL.RothwellE.E.GalyovP.A.BarrowJ.BurnsideP.WigleyDifferential cytokine expression in avian cells in response to invasion by Salmonella typhimurium, Salmonella enteritidis and Salmonella gallinarumMicrobiology146Pt 12200032173226
- S.KumarJ.J.BuzaS.C.BurgessGenotype-dependent tumor regression in Marek’s disease mediated at the level of tumor immunityCancer Microenviron220092331
- M.H.KogutL.RothwellP.KaiserPriming by recombinant chicken interleukin-2 induces selective expression of IL-8 and IL-18 mRNA in chicken heterophils during receptor-mediated phagocytosis of opsonized and nonopsonized Salmonella enterica serovar enteritidisMol Immunol402003603610
- I.EldaghayesL.RothwellA.WilliamsD.WithersS.BaluF.DavisonP.KaiserInfectious bursal disease virus: strains that differ in virulence differentially modulate the innate immune response to infection in the chicken bursaViral Immunol1920068391
- M.J.SlomkaV.J.CowardJ.BanksB.Z.LondtI.H.BrownJ.VoermansG.KochK.J.HandbergP.H.JorgensenM.Cherbonnel-PansartV.JestinG.CattoliI.CapuaA.EjdersundP.ThorenG.CzifraIdentification of sensitive and specific avian influenza polymerase chain reaction methods through blind ring trials organized in the European UnionAvian Dis512007227234
- K.J.LivakT.D.SchmittgenAnalysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) methodMethods252001402408
- S.FukuyamaY.KawaokaThe pathogenesis of influenza virus infections: the contributions of virus and host factorsCurr Opin Immunol232011481486
- H.M.YassineC.W.LeeR.GourapuraY.M.SaifInterspecies and intraspecies transmission of influenza A viruses: viral, host and environmental factorsAnim Health Res Rev1120105372
- M.C.ChanC.Y.CheungW.H.ChuiS.W.TsaoJ.M.NichollsY.O.ChanR.W.ChanH.T.LongL.L.PoonY.GuanJ.S.PeirisProinflammatory cytokine responses induced by influenza A (H5N1) viruses in primary human alveolar and bronchial epithelial cellsRespir Res62005135
- K.SuzukiH.OkadaT.ItohT.TadaM.MaseK.NakamuraM.KuboK.TsukamotoAssociation of increased pathogenicity of Asian H5N1 highly pathogenic avian influenza viruses in chickens with highly efficient viral replication accompanied by early destruction of innate immune responsesJ Virol83200974757486
- C.WatanabeY.UchidaH.ItoT.ItoT.SaitoHost immune-related gene responses against highly pathogenic avian influenza virus infection in vitro differ among chicken cell lines established from different organsVet Immunol Immunopathol1442011187199
- M.LinigerH.R.MoulinY.SakodaN.RuggliA.SummerfieldHighly pathogenic avian influenza virus H5N1 controls type I IFN induction in chicken macrophage HD-11 cells: a polygenic trait that involves NS1 and the polymerase complexVirol J920127
- A.N.CauthenD.E.SwayneM.J.SekellickP.I.MarcusD.L.SuarezAmelioration of influenza virus pathogenesis in chickens attributed to the enhanced interferon-inducing capacity of a virus with a truncated NS1 geneJ Virol81200718381847
- E.D.de GeusC.A.JansenL.VerveldeUptake of particulate antigens in a nonmammalian lung: phenotypic and functional characterization of avian respiratory phagocytes using bacterial or viral antigensJ Immunol188201245164526
- P.KaiserAdvances in avian immunology–prospects for disease control: a reviewAvian Path392010309324
- Biron CA, Sen GC. Innate responses to viral infections. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA (Eds.), Fields Virology. 5th ed., Philadelphia, USA: Lippincott Williams & Wilkins; 2007. p. 249–78.
- J.FriesenhagenY.BoergelingE.HrinciusS.LudwigJ.RothD.ViemannHighly pathogenic avian influenza viruses inhibit effective immune responses of human blood-derived macrophagesJ Leukoc Biol9220121120
