Abstract
Newcastle disease (ND) remains an important enzootic disease in chickens in several parts of the world. With the increasing reports of virulence and genetic diversity of the causative agent; Newcastle disease virus (NDV), there is a need to identify the circulating NDV in specific regions. In Oman, to this moment, such information is still lacking. The aim of this study was to isolate and characterize the NDV from ND outbreaks from commercial farms in Oman. Following suspected outbreaks of ND in three commercial farms in 2017, a total of 30 carcasses (10 from each flock) of adult chickens were subjected to necropsy for gross and histopathological examination, virus isolation and molecular methods. Specifically, haemagglutination inhibition (HI) test and reverse transcription-polymerase chain reaction (RT-PCR) assay were used for the virus detection and confirmation, respectively. Lesions were suggestive of viscerotropic velogenic form of ND based on gross and histopathological examinations. Isolation of NDV was present in 4 cases and further confirmed by RT-PCR following the target of the partial fusion protein gene of the viral genome. The sequence of the partial fusion gene was determined and phylogenetic tree was constructed based on the partial length F gene of 4 Omani isolates and 65 previously published NDVs. The findings predicted that the Omani isolates had high homology (99%) with the isolate from Pakistan belonging to genotype VII. Subsequently, the isolated pathotype was identified as the virulent NDV. This study serves as a basic work for further research on the analysis and phenotyping of NDV in the Sultanate of Oman. Improved monitoring and surveillance of the disease is important for proper preventive measures.
Keywords:
Introduction
Newcastle disease is regarded as a major production limiting disease in poultry farming. The disease is a well-known cause of high mortality resulting in huge economic loss in the poultry industry [Citation1,Citation2]. The disease is caused by the virulent forms of the avian Paramyxovirus 1 (APMV-1) also referred to as Newcastle disease virus (NDV) [Citation1]. The causative agent belongs to the genus Avulavirus of the family Paramyxoviridae and order Mononegavirales [Citation3]. Host and strain variations were reported to effect the severity of the produced disease [Citation4]. Based on the virulence and clinical manifestations, the viruses can be categorized into various pathotypes; viscerotropic velogenic, neurotropic velogenic, mesogenic, lentogenic, and asymptomatic enteric forms [Citation5]. Accordingly, high mortality reaching 100% (per acute disease) results from the virulent strains of NDV [Citation6], while the mesogenic or lentogenic strains might induce severe respiratory disease in immunocompromised birds [Citation7].
NDV is a negative-sense, single-stranded RNA virus with a genome length of about 15.2 kb [Citation8]. The NDV genome contains six genes that encode six proteins which include the nucleocapsid protein, fusion protein, matrix protein, phosphoprotein, hemagglutinin-neuraminidase, large RNA-dependent polymerase protein [Citation9]. To predict the virulence and tissue tropism of the virus, the amino-acid composition of the F protein cleavage site is vital [Citation10]. In addition, the HN protein length is applied in grouping the virus into avirulent and virulent strains. For instance, the HN protein lengths of 571 amino acids were reported in virulent strains [Citation11], whereas about 616 amino acids were observed in lentogenic strains [Citation12]. Other techniques employed for the same purpose include mean death time (MDT) and intra-cerebral pathogenicity index (ICPI) test using 9- to 11-day-old SPF, embryonated chicken eggs and 1-day-old SPF chickens, respectively [Citation13].
Overall, the detection and pathotyping of NDV from avian isolates is crucial for better understanding of the epizootiology of the disease. This is even more important with the recent reports of increasing genetic diversity of the virus and its widespread distribution in non-poultry avian species and wild birds [Citation14,Citation15].
In Oman, vaccination of birds against ND is little practiced in backyard flocks and recent studies depicted high seroprevalence of NDV in chickens [Citation16,Citation17]. Additionally, outbreaks of the disease have been reported even in vaccinated commercial flocks [Citation17]. To this moment, the strains of NDV responsible for the outbreaks remain unknown in the country. Particularly, molecular investigation of the sub genotype of NDVs and the analysis of the relationship of Oman’s NDVs with other isolates from different parts of the world are not available. Therefore, this study was aimed at isolating and characterizing the NDV from outbreaks in commercial flocks in the Sultanate of Oman.
Material and methods
.1 Study design
The study was carried out in the Sultanate of Oman located on the west coast of Gulf of Oman. The samples were collected from three commercial poultry farms located in Adakhilah and Albatinah, following the outbreaks of ND. Characteristics of the sampled farms and the signalment following the ND outbreaks are shown in . Samples of infected and dead birds were collected as well as documentation of parameters such as flock size, age, breed, clinical signs, morbidity and mortality during the postmortem examination was done. As such, detailed necropsy was conducted for all of the sampled dead birds. The weight of the sampled birds ranged from 400 to 850 g. Infected tissues such as intestine, proventriculus, trachea, spleen, and lungs were collected after post-mortem examination. Thereafter, the samples were placed in ice packs, transported to the laboratory, and stored at −25 °C for further analysis. The samples were submitted to Animal health Research Center, Oman. Approval for the study was obtained from the Department of Animal Health, Ministry of Agriculture, Oman.
able 1 Characteristics of the sampled farms with recorded ND outbreaks, age and weight of birds and mortality rate.
.2 Virus isolation
Virus isolation was carried out as outlined by OIE [Citation18]. Healthy eggs were collected from chickens that were sero-negative for ND. The sampled tissues were homogenized as a 10% (w/v) suspension in phosphate buffer saline (PBS) containing streptomycin and penicillin at 2 mg/mL and 2000 IU/mL, respectively. The suspension was clarified by centrifugation at 2000 rpm for 15 min. Thereafter, the allantoic cavity of the embryonated chicken eggs (9- to 11-day-old) was inoculated with 0.1 mL of the supernatant and incubated at 37 °C for 6 days. Candling of the eggs was done twice daily and the dead embryos were chilled overnight at 4 °C. The allantoic fluid was collected from the live and dead embryos. Detection of the virus was conducted by the slide haemagglutination test using chicken red blood cells (1%) and by RT-PCR assay.
.3 Isolation of viral RNA, RT-PCR amplification and phylogenetic analysis
The viral RNA was extracted from the allantoic fluid by using the TRIzol LS reagent (Invitrogen, USA) following the manufacturer’s instructions. The reaction mixture comprised of 150 μL of allantoic fluid, 570 μL of VNE Buffer, 570 μL of ethanol, 500 μL Wash Buffer 1 and 750 μL of wash buffer. Each RNA sample was dissolved in 40 μL sterile RNAse-free water and stored at −70 °C. The same extracted RNA was used as template for RT-PCR amplification. The major steps carried out in RT-PCR for DNA amplification included synthesis of the complementary DNA (cDNA) by using the reverse transcription enzyme (48 °C for 30 min). This was followed by an initial denaturation (94 °C for 2 min), denaturation of 40 cycles (94 °C at 15 s), annealing (52 °C for 30 s), and extension (68 °C for 60 s). For the positive and negative controls, the viral RNA was extracted from a live ND vaccine and non-template control (water), respectively. Specific primer sequences used for the RT-PCR and amplification of the partial fusion gene comprises of 5′-GCAGCTGCAGGGATTGTGGT-3′ (forward) and 5′-TCTTTGAGCAGGAGGATGTTG-3′ (reverse) [Citation19]. The RT-PCR reaction was set up in a total volume of 50 µL per reaction comprised of 5.0 μL 10 × buffer, 2.0 μL 25 mM MgCl2, 2.0 μL 10 mM dNTP, 0.2 μL Taq, and 0.8 μL NDV-F. RT-PCR products were resolved in an agarose gel (1.5%) containing ethidium bromide and visualized under UV light.
Nucleotide sequence of the PCR products was determined using the forward and reverse primers. The raw data sequence was assessed using Basic Local Alignment Search Tool (BLAST) and compared to other sequences in the GenBank NCBI [Citation20]. To investigate the evolutionary associations, the fusion protein gene of the NDV isolates was compared to 65 nucleotide sequences reported from other studies (Supplementary Table 1). Using the ClutalW multiple alignment technique (MEGA 7 software), sequence and phylogenic analysis of the partial F and HN genes were conducted as described by Miller et al. [Citation21]. Construction of the phylogenetic tree was done by applying the maximum likelihood method whereas the criteria described by Diel et al. [Citation22] were used in classifying the isolated viruses.
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.ijvsm.2018.08.007.
Nucleotide sequence of the PCR products was determined using the forward and reverse primers. The raw data sequence was assessed using Basic Local Alignment Search Tool (BLAST) and compared to other sequences in the GenBank NCBI [Citation20]. To investigate the evolutionary associations, the fusion protein gene of the NDV isolates was compared to 65 nucleotide sequences reported from other studies (EquationSupplementary Table 1). Using the ClutalW multiple alignment technique (MEGA 7 software), sequence and phylogenic analysis of the partial F and HN genes were conducted as described by Miller et al. [Citation21]. Construction of the phylogenetic tree was done by applying the maximum likelihood method whereas the criteria described by Diel et al. [Citation22] were used in classifying the isolated viruses.
Results
In this study we focused on the isolation and characterization of the viruses from suspected ND outbreaks in three commercial poultry farms in Oman in 2017. This was based on both field virus isolation and characterization at molecular level. The mortality in infected chickens ranged from 14 to 63% following the onset of the clinical disease (). Necropsy examination revealed tracheitis with petechial hemorrhage in the proventriculus as well as splenic congestion. Using the allantoic route, nine-day-old embryonated SPF chicken eggs were inoculated with the suspension obtained from the pooled samples of the internal organs (lung, spleen, trachea, and brain) from the samples positive for NDV specific HI test. Based on the application of RT-PCR for viral isolation, all the four samples were confirmed as NDV.
Results revealed 356 bp products following the amplification of the partial fusion protein gene using RT-PCR. Excluding the genotype XV, all the sequences from all existing genotypes (I to XVII) were used to build the general tree to identify the genotype of the newly studied sequences. Phylogeny analysis of partial sequences of the selected strains partial fusion gene showed that the isolated viruses (OM ND 2/chicken/Oman/2017, OM ND 4/chicken/Oman/2017, OM ND 5F/Chicken/Oman/2017, and OM ND 6F/Chicken/Oman/2017) belonged to the genotype VII (). The isolated viruses displayed an average distance per site which was less than 10% (0.1) (Supplementary Table 2). Calculation of the mean intra-genotype genetic diversity using the mean genetic distance per site was less than 3% at the recommended bootstrap values (<60). The amino acids sequences of the fusion protein proteolytic cleavage site motifs (residues 112–117) were identical in the entire isolated virus. Following the multiple sequence alignment, our NDV isolates revealed about 99% phylogenetic similarity to the Pakistan isolate based on the nucleotide and amino acids identities.
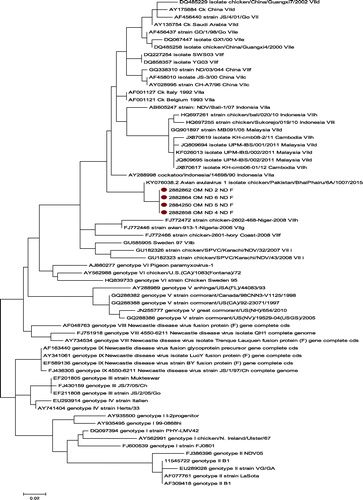
Results revealed 356 bp products following the amplification of the partial fusion protein gene using RT-PCR. Excluding the genotype XV, all the sequences from all existing genotypes (I to XVII) were used to build the general tree to identify the genotype of the newly studied sequences. Phylogeny analysis of partial sequences of the selected strains partial fusion gene showed that the isolated viruses (OM ND 2/chicken/Oman/2017, OM ND 4/chicken/Oman/2017, OM ND 5F/Chicken/Oman/2017, and OM ND 6F/Chicken/Oman/2017) belonged to the genotype VII (). The isolated viruses displayed an average distance per site which was less than 10% (0.1) (EquationSupplementary Table 2). Calculation of the mean intra-genotype genetic diversity using the mean genetic distance per site was less than 3% at the recommended bootstrap values (<60). The amino acids sequences of the fusion protein proteolytic cleavage site motifs (residues 112–117) were identical in the entire isolated virus. Following the multiple sequence alignment, our NDV isolates revealed about 99% phylogenetic similarity to the Pakistan isolate based on the nucleotide and amino acids identities.
Discussion
Detection and pathotyping of NDV is vital in understanding the epizootiology of the virus in any region. This is more important with the growing need for the evaluation of the efficacy of existing ND vaccines [Citation5]. In Oman, ND has been identified as a major cause of setbacks in the poultry industry [Citation16,Citation17]. In this study, we were able to isolate and characterize the NDV responsible for the outbreaks recorded in three large commercial flocks in Oman.
Isolation of the virus was achieved by inoculation of the suspension obtained from the specific organs of the affected birds into embryonated eggs of SPF chicken (9–11 day old). Also, clinical samples were positive for NDV following the use of RT-PCR. This technique has been used by other authors for the same purpose [Citation12]. The isolates in this study induced embryonic death within 60 h post-inoculation, thus depicting the virulent nature of the virus. There were variable mortalities (12–63%) amongst the investigated broiler chickens. Also, there were NDV infection lesions such as tracheitis and proventriculus haemorrhage which are characteristic features of velogenic NDV [Citation2,Citation13].
Based on the F protein phylogeny, the isolated viruses in this study were all related to the velogenic NDV strains, shared a monophyletic branch with the Pakistani isolate (KY076038.2 Avian avulavirus 1 isolate chicken/Pakistan/BhaiPhairu/6A/1007/2015), and subsequently classified as genotype VII. These groups of viruses belong to the class II, genetically diverse, and suggested to have emerged from the Far East before spreading to other parts of the world [Citation1,Citation23]. Molecular pathotyping has been recognised as more rapid and reliable technique for NDV pathotyping as compared to MDT and ICPI tests [Citation24,Citation25]. Hence, in this study, the method was applied to the NDV strains based on their amino-acid sequences of the fusion protein proteolytic cleavage site motifs (residues 112–117). Accordingly, the cleavage site motif 112RRQKRF117 known as a characteristic for vNDV was shared by all the studied strains. Similar result was obtained following the molecular pathotyping of NDV strains recently isolated from broilers in Egypt [Citation25].
Presently, the genotype VII viruses are grouped into nine various sub-genotypes (VIIa-VIIi), reoccurring in the Middle East and Asia [Citation1]. All of them are predicted to be virulent according to deduced amino acid motifs [Citation1,Citation21]. Furthermore, the calculated mean intra-genotype genetic diversity using the mean genetic distance/site was <3% at the recommended bootstrap values (<60). This method was described by Diel et al. [Citation22] as the required cut-off in assigning a new sub-genotype. Based on the latter criteria, all the studied isolates were classified as member of the sub-genotype VIIi.
Recent studies have shown that the lately identified viruses (sub-genotype VIIi) are circulating between the Middle East and Southeast Asia, with potential of fast spread to Eastern Europe and North Africa [Citation1,Citation26]. Dimitrov et al. [Citation1] also suggested that the sub-genotype VIIi to have emerged recently, with their primary isolation from chickens in Pakistan, Israel and Indonesia. In this study, the isolates were obtained from vaccinated commercial farms, which is consistent with the findings obtained in Pakistan [Citation26]. Also, pheasants in the latter country during the 2011–2012, recorded high case fatality rates as well as economic losses in susceptible birds following vNDV infection caused by the same sub-genotype VIIi [Citation27,Citation28]. These findings suggest panzootic potential of the viruses based on the previous reports in vaccinated birds and their recent emergence and downward spread from Indonesia to Pakistan [Citation21], and now to Oman.
ND is regarded as an enzootic disease in Oman [Citation16,Citation17]. The findings herein with reports of vNDV presence in vaccinated birds could imply increasing replication of the virus, thus precipitating high environmental viral load. Such event is enhanced by the constant evolution of the viruses [Citation29]. Unvaccinated birds, especially the backyard chickens are predisposed to high mortalities following the infection by these vNDV [Citation21]. In Oman, backyard poultry remains a significant practice and lack of vaccination was found as a risk factor for the high seroprevalence to NDV antibodies [Citation17]. Such event might contribute to the continuous circulation of the virus between backyard and commercial flocks. For instance, backyard flocks or non-poultry birds are often found surrounding large commercial farms lacking adequate biosecurity [Citation1]. In Pakistan, non-poultry birds in close proximity to poultry birds were found to play an important role in the circulation of NDV in the country [Citation14]. However, such event need to be equally investigated in Oman based on the similarity between the isolated viruses.
In Oman, there is widespread use of vaccine strains including Lasota, B1 and F for the prevention and control of ND infection. Nevertheless, this study showed that virulent strains of NDV are still present in vaccinated flocks. Existing vaccines have shown the potential of reducing virus replication [Citation5]. The fact that backyard flocks in the country are rarely vaccinated [Citation30] means that NDV remains a strong threat to the poultry industry. These practices especially issues with biosecurity could be vital for the genetic similarity between the NDV isolates identified in Oman in the present study, with those of Pakistan. Molia and coworkers explained that effective quarantine and disease reporting system are essential in the prevention of ND [Citation31]. Hence, the results from this study indicate the need for proper monitoring and surveillance for economically important transboundary diseases like ND in Oman.
Conclusions
The current study is the first attempt to isolate and characterize the NDV field strains in chickens from the Sultanate of Oman. Results herein showed that the circulating NDV in Oman is the virulent NDV and it is responsible for the outbreaks recorded in the sampled commercial flocks. Also, phylogenetic analysis showed that the virus is similar to the Pakistan isolate. These findings call for optimum transboundary monitoring and surveillance for proper control of the disease. This study provides the basis for further research on the analysis and phenotyping of NDV in the country. Continuous surveillance of the disease is important to advance our understanding of ND epizootiology in chickens in Oman.
Acknowledgements
Our appreciation goes to the staff of Directorate General for Animal Wealth, Reference laboratory, and the Department of Veterinary Services in the Sultanate of Oman, for their cooperation and understanding.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interests
The authors state that they have no competing interest regarding this manuscript.
Notes
Peer review under responsibility of Faculty of Veterinary Medicine, Cairo University.
References
- K.M.DimitrovA.M.RameyX.QiuJ.BahlC.L.AfonsoTemporal, geographic, and host distribution of avian paramyxovirus 1 (Newcastle disease virus)Infect Genet Evol3920162234
- M.B.SadiqB.R.MohammedThe economic impact of some important viral diseases affecting the poultry industry in Abuja, NigeriaSok J Vet Sci152017717
- G.K.AmarasingheN.G.Aréchiga CeballosA.C.BanyardC.F.BaslerS.BavariA.J.BenettTaxonomy of the order: Mononegavirales: update 2018Arch Virol163201822832294
- C.W.BeardR.P.HansonNewcastle diseaseM.S.HofstadH.J.BarnesB.W.CalnekW.M.ReidH.W.YoderDisease of Poultry8th ed.1984Iowa state University PressAmes, IA452470
- K.M.DimitrovC.L.AfonsoQ.YuP.J.MillerNewcastle disease vaccines-A solved problem or a continuous challenge?Vet Microbiol2062017126136
- OIE. World Organization for Animal Health. In: Manual of Diagnostic Tests and Vaccines for Terrestrial animals. 7th ed. Ch. 2.3.14. OIE, Paris, France; 2012. p. 556–73.
- Lamb RA, Collins PL, Kolakofsky D, Melero JA, Nagai Y, Fauquet CM. Family Paramyxoviridae. Virus taxonomy: The classification and nomenclature of viruses. The eighth report of the international committee in taxonomy of viruses. Fauquet CM (Ed.); 2005.
- E.W.AldousJ.K.MynnJ.BanksD.J.AlexanderA molecular epidemiological study of avian paramyxovirus type 1 (Newcastle disease virus) isolates by phylogenetic analysis of a partial nucleotide sequence of the fusion protein geneAvian Pathol322003239256
- D.J.AlexanderJ.G.BellR.G.AldersFAO Technology Review: Newcastle disease with special emphasis on its effect on village chickensFAO Anim Prod Health4ISSN 0254-6019200455
- Y.WangZ.JiangZ.JinH.TanB.XuRisk factors for infectious diseases in backyard poultry farms in the Poyang Lake area, ChinaPLoS One82013e67366
- P.YuanR.G.PatersonG.P.LeserR.A.LambT.S.JardetzkyStructure of the ulster strain newcastle disease virus hemagglutinin-neuraminidase reveals auto-inhibitory interactions associated with low virulencePLoS Pathog82012115
- V.S.DhaygudeG.K.SawaleM.M.ChawakN.R.BulbuleS.D.MoregaonkarD.S.GavhaneMolecular characterization of velogenic viscerotropic Ranikhet (Newcastle) disease virus from different outbreaks in desi chickensVet World102017319323
- G.CattoliA.FusaroI.MonneS.MoliaA.Le MenachB.MaregeyaEmergence of a new genetic lineage of Newcastle disease virus in West and Central Africa-Implications for diagnosis and controlVet Microbiol1422011168176
- A.WajidK.M.DimitrovM.WasimS.F.RehmaniA.BasharatT.BibiReported isolation of virulent Newcastle disease viruses in poultry and captive non-poultry avian species birds in Pakistan from 2011 to 2016Prev Vet Med142201716
- A.J.AyalaK.M.DimitrovC.R.BeckerI.V.GoraichukC.W.ArnsV.I.BolotinPresence of vaccine-derived Newcastle disease viruses in wild birdPLoS ONE112016e0162484
- T.A.L.ShekailiH.CloughK.GanapathyM.BaylisSero-surveillance and risk factors for avian influenza and Newcastle disease virus in backyard poultry in OmanPrev Vet Med1222015145153
- A.AlsahamiA.IderisA.OmarS.Z.RamanoonM.B.SadiqSeroprevalence of newcastle disease virus in backyard chickens and herd-level risk factors of newcastle disease in poultry farms in OmanInt J Vet Sci Med2018 10.1016/j.ijvsm.2018.06.004
- O.I.E.Manualof Diagnostic tests and vaccines for terrestrial animalsWorld Organisation for Animal Health201311851191
- T.NanthakumarR.S.KatariaA.K.TiwariG.ButchaiahJ.M.KatariaPathotyping of Newcastle disease viruses by RT-PCR and restriction enzyme analysisVet Res Commun242000275286
- S.KumarG.StecherK.TamuraMEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasetsMol Biol Evol33201618701874
- P.J.MillerR.HaddasL.SimanovA.LublinS.F.RehmaniA.WajidIdentification of new sub-genotypes of virulent Newcastle disease virus with potential panzootic featuresInfect Genet Evol292015216229
- D.G.DielL.H.da SilvaH.LiuZ.WangP.J.MillerC.L.AlfonsoGenetic diversity of avian paramyxovirus type 1: proposal for a unified nomenclature and classification system of Newcastle disease virus genotypesInfect Genet Evol12201217701779
- T.A.KhanC.A.RueS.F.RehmaniA.AhmedJ.L.WasilenkoP.J.MillerPhylogenetic and biological characterization of Newcastle disease virus isolates from PakistanJ Clin Microbiol48201018921894
- K.GanarM.DasS.SinhaS.KumarNewcastle disease virus: current status and our understandingVirus Res18420147181
- S.S.EweisA.AliS.M.TamamM.H.MadboulyMolecular characterization of Newcastle disease virus (genotype VII) from broiler chickens in EgyptBeni-Suef University J Basic Appl Sci62017232237
- S.F.RehmaniA.WajidT.BibiB.NazirN.MukhtarA.HussainPresence of virulent Newcastle disease virus in vaccinated chickens in farms in PakistanJ Clin Microbiol53201517151718
- M.MunirM.CorteyM.AbbasZ.U.QureshiF.AfzalM.Z.ShabbirBiological characterization and phylogenetic analysis of a novel genetic group of Newcastle disease virus isolated from outbreaks in commercial poultry and from backyard poultry flocks in PakistanInfect Genet Evol12201210101019
- M.Z.ShabbirS.ZohariT.YaqubJ.NazirM.A.B.ShabbirN.MukhtarGenetic diversity of Newcastle disease virus in Pakistan: a countrywide perspectiveVirol J102013170
- P.J.MillerL.M.KimH.S.IpC.L.AfonsoEvolutionary dynamics of Newcastle disease virusVirology39120096472
- B.Al-QamashouiO.MahgoubI.KadimE.SchlechtTowards conservation of Omani local chicken: phenotypic characteristics, management practices and performance traitsAsian-Australas J Anim Sci272014767777
- S.MoliaI.A.BolyR.DubozB.CoulibalyJ.GuitianV.GrosboisLive bird markets characterization and trading network analysis in Mali: Implications for the surveillance and control of avian influenza and Newcastle diseaseActa Trop15520167788
