Abstract
To isolate fungal communities from coastal areas of the Red Sea in Saudi Arabia and identify and classify them by molecular techniques. Samples were collected from the seaside of the Red Sea in Jeddah, Saudi Arabia during March 2012 and stored in sterile screw cap bottles for further analysis. Phenotypic and genotypic characterization of fungal isolates were done using standard techniques. Eight fungal genera including Aspergillus, Penicillium, Thielavia, Fusarium, Emericella, Cladosporium, Scytalidium and Alternaria. Most isolated fungi showed significant growth on petroleum media and were thus considered capable of biodegradation of crude oil based substances. The fungal genera isolated from the Red Sea had 97–100% similarity with the related fungi recorded in the GenBank in which they were deposited. The morphological and molecular structure of these marine fungal isolates closely resembles their terrestrial counterparts in the GenBank. The capabilities of these fungal species to utilize petroleum as a source of carbon speaks to future applications in which marine fungi may be utilized in the breakdown of petroleum-based waste in an ecologically efficient manner.
1 Introduction
Fungi are fundamental organisms in our ecological systems and will be found anywhere where the environment will support their growth, both on land and in water (CitationPang and Mitchell Julian, 2005). However, our understanding of fungal diversity particularly in marine environment is still scarce compared to our knowledge of bacteria and viruses in such environments. Most of the information that we have is based on research that was done in the laboratory, for this reason a selective group of very few fungal genera are being studied rigorously (CitationAnderson and Cairney, 2004). In recent years, there has been a growing interest in the enormous biodiversity of marine fungal communities, their significant contribution to the production of useful bioactive compounds and their contribution to equilibrium within marine environments (Antunes et al., Citation2011; Masuma et al., Citation2001).
Fungal communities exist and thrive in the sea. It had long been assumed that bacteria are the most prevalent saprophytic species in marine environments and the largest contributors to organic matter decomposing, however many fungi also play major roles in such environments. Aquatic Fungi may thrive as obligate marine species (that grow and sporulate exclusively and permanently in seawater) or as facultative marine fungi (those that grow in freshwater milieu as well as seawater). Terrestrial fungi can also grow within marine environments when conditions are suitable (Hyde et al., Citation1998; Jones and Pang, Citation2012). Earlier studies on marine fungi in the Red Sea and other briny bodies of water have recognized the prevalence of the following fungal species: Ascomycota, Basidiomycota and mitosporic fungi (CitationSimões et al., 2015). Other studies also found Candida, Cryptococcus, Debaryomyces and Rhodotorula prevalent in seawater (CitationAbd-Elaah, 1998). It has been confirmed that the extreme conditions conferred by the environment of the Red Sea and other similarly hostile environments such as the black sea have enabled eukaryotes especially fungi to substantially evolve and develop stress-resistant mechanisms to cope with their new ecological niches (CitationWang et al., 2014). Fungal communities in the Red Sea studies also revealed the presence of terrestrial fungal species including Aspergillus sp., Penicillium sp. and Fusarium, Neurospora and Rhizopus (Abd-Elaah, Citation1998; Basem et al., Citation2012). We conducted a similar study (published in 2013) to quantify and classify the terrestrial fungal species off the coast of Jeddah, Saudi Arabia and found that Aspergillus and Penicillium were to most common terrestrial species in the isolated from the region (CitationAlwakeel, 2013).
Interestingly, terrestrial fungi such as Aspergillus have been shown to make many adaptations when exposed to hypersaline environments such as seawater. Aspergillus had been shown to produce antimicrobial alkaloids that limit hyphal growth when exposed to saltwater (CitationMasuma et al., 2001). The marine-derived fungus Curvularia has also been reported to produce a novel antimicrobial alkaloid when exposed to the correct environmental pressures (CitationYang et al., 2016). Such observations contribute to the growing role of marine fungi in aquatic environments as not only saprophytic organisms but as possible contributors to reduced pollution and maintainers/regulators of prokaryotic populations in the sea (CitationWang et al., 2014). A growing body of research had found that soil fungi species are capable of breaking down kerosene at various concentrations and utilizing it as a source of carbon but very few studies had examined this capacity in marine fungi (CitationSimister et al., 2015).
The Kingdom of Saudi Arabia relies heavily on the desalinization of seawater as a source of drinking water through desalination plantations located in most of the coastal regions of the kingdom. Because of this, there is a need to determine and maintain high standards when it comes to the safety of using seawater for drinking purposes. Although there is much contact with the organisms from seawater in the waters of the Red Sea, very few studies had attempted to classify and characterize Red Sea fungal species and the chemical nature of their metabolites. Hence the aim of this study to identify, both morphologically and molecularly, the fungal species occurring in the Red Sea coastal areas of Jeddah, Saudi Arabia in an effort to contribute to the taxidermy of fungi in the region and to identify the capacity of these isolates as petroleum biodegraders.
2 Methods
2.1 Sampling technique
Seventy-seven samples consisting of seawater and sand were collected from the seaside area of Masturah (latitude: 23°5′31.46″ N/longitude: 38° 49′17.52″ E) near Jeddah, Saudi Arabia in March 2012. Each sample consisted of both water and sandy soil. Samples were collected in sterile 300 ml screw cap bottles from an area measuring approximately 37 km2 of the Red Sea. The region in question is port for small boats as well as a known fishing area. In order to obtain a representative sample, we collected samples from different depths. In areas near the shoreline where the water was shallow, samples were collected by manually filling the bottles with sand and seawater. In deeper areas, the bottles were lowered into the sea to acquire the samples. Collected samples were stored in a refrigerator until they were processed no longer than 24 h later.
2.2 Fungal isolation
2.2.1 Sand components of the samples
Ten grams of each sand sample was diluted in 90 ml of sterile distilled water. 0.5 of this suspension was spread, in triplicates, on glucose-Czapek’s agar (for isolation and enumeration purposes) and petroleum oil-Czapek’s agar for assessment of fungal biodegradation capacities (see below). The plates were incubated for at 28 °C and colony growth was observed 3–4 days later. The complete incubation period was 7 days. As soon as colonies appeared, they were transferred to test tubes containing Sabouraud agar media diluted in sterile sea water in order to confirm the colony’s purity and maintain the osmolality. Colonies were thus sub-cultured in glass tubes. Colonies were identified phenotypically based macroscopic and microscopic appearance.
2.2.2 Seawater potion of the sample
Ten milliliters of each sample were diluted in 90 ml of sterile distilled water to obtain a dilution of 1/10 M. one milliliter of the previously prepared 1/10 M dilution was further diluted in 9 ml of sterile distilled water to achieve a dilution of 1/100 M. One milliliter of each of the prepared 1/100 M dilutions was spread on Sabouraud agar plates and incubated at 28 °C for 7 days. Three replicas were made for each dilution. Fungal colonies were later counted and identified morphologically.
2.3 Petroleum (i.e. crude oil) Biodegradation Potential
In order to determine the ability of the fungi within the samples to utilize crude oil as carbon source we used a special preparation of the Czapek Dox Broth with a modification. The Czapek media is composed of 30 g/L of sucrose as a carbon source. Instead of using sucrose we used 10 ml/L of petroleum to thus prepare petroleum oil-Czapek’s agar. In order to maintain the osmolality of the environment from which the samples were obtained, sterilized salt water was added to the mixture. The samples were then incubated at 28 °C for 7 days. Fungal mycelia extension on the plate (i.e. colony diameter) as well as colony numbers after 7 days were assessed and used to express an isolate’s ability to utilize petroleum as a source of carbon.
2.4 Phenotypic characterization
All isolates grew equally well on Czapek’s dextrose agar plates at 28 °C. Most colonies grew up to 3–4 cm in diameter after 48 h of incubation. According to their growth characteristics and colony morphology, 16 well-isolated colonies were selected at random for further characterization and genetic identification.
2.5 Genotypic sequencing identification of fungal isolates
A small amount of fungal growths was scraped and suspended in 100 μl sterile distilled water in 2 ml sterile vials and boiled at 100 °C for 15 min. Fungal DNA was extracted and isolated using SolGent purification bead. The ribosomal rRNA gene was amplified using universal fungal primers: internal transcribed spacer 1 (ITS1: 5′-TCC GTA GGT GAA CCT GCG G-3′), and internal transcribed spacer 4 (ITS4: 5′-TCC TCC GCT TAT TGA TAT GC-3′). The PCR reaction mixture was prepared using Solgent EF-Taq as follows: 10× EF-Taq buffer 2.5 μl, 10 mM dNTP (T) 0.5 μl, primer (Forward-10 pmol) 1.0 μl, primer (Reverse-10 pmol) 1.0 μl, EF-Taq polymerase (2.5 U) 0.25 μl, DNA template 1.0 μl, Distilled Water (up to 25 μl). Then the amplification was carried out in a thermal cycler under the following conditions: one round of denaturation at 95 °C for 15 s followed by 30 cycles of denaturation at 95 °C for 20 s, annealing at 50 °C for 40 s and extension at 72 °C for 1 min, with a final extension step at 72 °C for 5 min. The PCR products were then purified with the SolGent PCR Purification Kit-Ultra (SolGent, Daejeon, South Korea). By running the products in a 1% agarose gel by electrophoresis, molecular sizes of the purified PCR products were identified. Then these bands were eluted and sequenced with the incorporation of dideoxynucleotides (dd NTPs) in the reaction mixture. Each sample was sequenced in the sense and antisense directions using ITS1 and ITS4 primers to ensure the accuracy of the nucleotide sequences. The ITS sequences obtained were further analyzed using BLAST search program from the National Center of Biotechnology Information (NCBI) website to find identical species in the GenBank®. The methodology described closely resembles methods of rapid fungal species identification described by Turenne et al. (CitationTurenne et al., 1999). Phylogenetic data described below were obtained by alignment and phylogenetic analysis of the fungal sequences. The nucleotide sequences were aligned by using the MegAlign®. Constructing the phylogenic tree was done via neighbor-joining analysis (CitationSaitou and Nei, 1987) the DNASTAR computer program (Version 5.05. DNASTAR. Madison, WI).
3 Results and discussion
Of the 77 samples inoculated on petroleum oil-Czapek’s agar pates, 34 yielded no fungal growth after incubation and 43 samples grew fungi. shows the total colony counts, frequencies and percentages of genera and species that grew on the petroleum oil-Czapek’s agar. Of all the isolates, Penicillium chrysogenum produced the greatest number of colonies per plate (a total colony count of 45 CFU) thus displaying the greatest capacity to break down petroleum. However, P. chrysogenum was only found in 2 samples. The most prevalent species to grow on petroleum oil-Czapek’s media was Aspergillus flavus which grew on a total of 14 samples of the total 43 samples examined, followed by Aspergillus niger (found in 11 samples) then Aspergillus terreus (found in 8 samples).
Table 1 Fungal isolates total colonies, frequency per whole sample and percentage of growth on petroleum oil-Czapek’s agar.
Random selection of fungal isolates revealed a total of 8 fungal genera (totaling to 14 fungal species), which later underwent molecular analysis. These include the following fungal genera and species:
| (1) | Aspergillus – 5 species (caespitosus, fumigatus, ochraceus, sydowii and terreus) | ||||
| (2) | Penicillium – 3 species (chrysogenum – 2 isolates, citrinum, oxalicum) | ||||
| (3) | Thielavia hyalocarpa | ||||
| (4) | Fusarium thapsinum (Teleomorph: Gibberella thapsina) | ||||
| (5) | Emericella nidulans (4 isolates) | ||||
| (6) | Cladosporium cladosporioides | ||||
| (7) | Scytalidium hyalinum | ||||
| (8) | Alternaria brassicae | ||||
The amplified fungal sequences were used as BLAST queries against the NCBI database. shows the fungi isolated from the Red Sea compared with closely related fungi in the GenBank®. Most of the fungi we isolated had 97–99% similarity with the related fungi recorded in the bank. Nucleotide sequences of the ITS1 region of 18s rRNA genes of isolated fungal species were deposited in GenBank database under accession numbers listed in below.
Table 2 Identification of fungal isolates recovered from Red Sea’s coast in Jeddah, Saudi Arabia the by sequencing of internal transcribed spacer 1 and 4 (ITS1 and ITS 4) and region of rRNA gene compared with sequences listed in the GenBank for similar species.
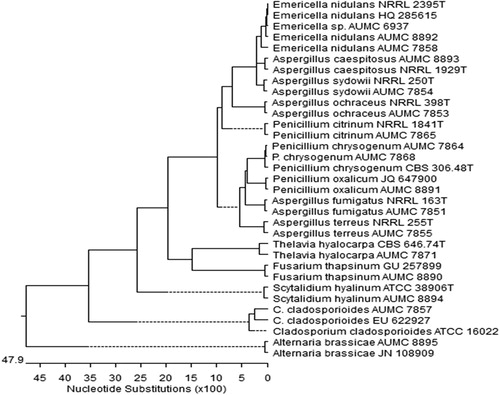
shows the phylogenetic tree of the fungal species that were isolated from Red Sea water sediments. The sequences of the PCR amplicons were found to be 97–100% similar to the sequences of 18S rRNA regions of the respective genera and species of closely related fungi documented in the GenBank®. These finding allow us to infer homology from the degree of similarity found between our isolated and those within the GenBank® database, which we concluded, were very similar.
Figure 1 Phylogenetic tree for fungal species isolated from Red Sea water sediments (given the AUMC Numbers), P = Penicillium and C = Cladosporium. The scale indicates the number of nucleotide substitutions per site. Reference type strains of corresponding fungi are involved in the tree (given CBS, ATCC, JN, JQ, HQ, GU, NRRL numbers).
The aim of this study was to identify, both macroscopically and molecularly, the most prevalent fungal species in the Jeddah Red Sea coast and to assess the capacity of these species to grow on media inoculated with petroleum thus establishing whether or not fungi can utilize crude oil as a source of energy.
Recent oil spills in bodies of water such as the Gulf of Mexico raise concerns over the possibility of utilizing naturally occurring organisms in the biodegradation and biomediation of such waste products. We have thus embarked on investigating the marine biodiversity in Saudi coast of the Red Sea and assessing their capabilities of growth on media that contains petroleum in an effort to mimic a crude oil spill in a body of saltwater. In this study, we identified several fungal communities from the sand and seawater in the Red Sea coastline in Jeddah, Saudi Arabia. Fungal isolates included Aspergillus, Penicillium and fusarium genera among many others. Such species have been reported previously as both terrestrial and facultative marine fungi in the literature (Antunes et al., Citation2011; Pecoraro et al., Citation2015). Many studies in the field have aimed to isolate fungi from floating debris and mangrove thus yielding species such as Swampomyces triseptatus which readily grow on plants and driftwood but may be harder to come by when isolating samples of seawater and sand (CitationAbdel-Wahab et al., 2014). The ability of the fungal isolates found, particularly Aspergillus, to survive within hypersaline environments indicates its high resilience and ability to adapt (CitationKis-Papo et al., 2003).
Comparison of the fungal isolates with those within the GenBank® has revealed a high degree of homology between those within the bank. This indicates that similar terrestrial isolates are known to exist and that a small degree of evolution had occurred in the species in order to promote their survival. This degree of homology with previous isolates also indicates that the fungal species have not been exposed to factors, environmental or otherwise that would promote further genetic diversity i.e. what has been frequently referred to as concerted evolution (CitationGanley and Kobayashi, 2007). Similar species to the ones isolated have been reportedly found in isolates from other briny bodies of water in the area, such as the Mediterranean and the Dead Sea (Kis-Papo et al., Citation2003; Pecoraro et al., Citation2015). The overall capacity of phylogenetically closely-related fungal species to utilize petroleum as a source of energy is further evidence that these species had and continue to be exposed to petroleum and have thus developed the evolutionary means by which to degrade it and utilize it.
The capacity of growth on petroleum media displayed by A. flavus and A. sydowii confirm the concerns of possibly utilizing these fungi as agents of biodegradation or biomediation. Such biological capacities should be further investigated in order to find the exact biochemical pathway involved in crude oil degradation for future environmental and industrial applications.
4 Conclusion
In conclusion, we are just beginning to understand the diversity of Marine fungal species of the Red Sea as an ecological entity. We have found various species of fungi that resemble both morphologically and molecularly their terrestrial counterparts. Many of the isolates showed good growth potential on media containing crude oil thus demonstrating their capacity as possible naturally occurring biodegrading agents. Further studies are needed to elucidate the exact mechanisms by which these fungi break down petroleum.
Conflict of interest
There were no conflicts of interest in the conduction of study.
Notes
Peer review under responsibility of University of Bahrain.
References
- G.A.Abd-ElaahThe occurrence of fungi along the Red Sea coast and variability among isolates of Fusarium as revealed by isozyme analysisJ. Basic Microbiol.385–6199830331110.1002/(SICI)1521-4028(199811)38:5/6<303::AID-JOBM303>3.0.CO;2-E
- M.A.Abdel-WahabM.S.HodhodA.H.A.BahkaliE.B.JonesMarine fungi of Saudi ArabiaBot. Mar.5742014323335
- S.S.AlwakeelIsolation and identification of fungal and bacterial specimens from the sand and seawater of the red sea coastline of Saudi ArabiaAdv. Environ. Biol.201313661374
- I.C.AndersonJ.W.G.CairneyDiversity and ecology of soil fungal communities: increased understanding through the application of molecular techniquesEnviron. Microbiol.68200476977910.1111/j.1462-2920.2004.00675.x
- A.AntunesD.K.NgugiU.StinglMicrobiology of the Red Sea (and other) deep-sea anoxic brine lakesEnviron. Microbiol. Rep.34201141643310.1111/j.1758-2229.2011.00264.x
- M.J.BasemA.-S.RolaA.-N.TariqIsolation and molecular identification of Ascomycetes in sediments and waters of the Gulf of Aqaba, Red SeaNat. Sci.2012
- A.R.D.GanleyT.KobayashiHighly efficient concerted evolution in the ribosomal DNA repeats: total rDNA repeat variation revealed by whole-genome shotgun sequence dataGenome Res.172200718419110.1101/gr.5457707
- K.D.HydeE.B.G.JonesE.LeañoS.B.PointingA.D.PoonythL.L.P.VrijmoedRole of fungi in marine ecosystemsBiodivers. Conserv.7919981147116110.1023/a:1008823515157
- E.B.G.JonesK.L.PangMarine fungi: and fungal-like organisms2012De Gruyter
- T.Kis-PapoA.OrenS.P.WasserE.NevoSurvival of filamentous fungi in hypersaline Dead Sea waterMicrob. Ecol.452200318319010.1007/s00248-002-3006-8
- R.MasumaY.YamaguchiM.NoumiS.OmuraM.NamikoshiEffect of sea water concentration on hyphal growth and antimicrobial metabolite production in marine fungiMycoscience425200145545910.1007/BF02464342
- K.-L.PangI.Mitchell JulianMolecular approaches for assessing fungal diversity in marine substrataBot. Mar.482005332
- L.PecoraroM.GirlandaZ.-J.LiuL.HuangS.PerottoMolecular analysis of fungi associated with the Mediterranean orchid Ophrys bertolonii MorAnn. Microbiol.65420152001200710.1007/s13213-015-1038-9
- N.SaitouM.NeiThe neighbor-joining method: a new method for reconstructing phylogenetic treesMol. Biol. Evol.441987406425
- R.L.SimisterC.M.PoutasseA.M.ThurstonJ.L.ReeveM.C.BakerH.K.WhiteDegradation of oil by fungi isolated from Gulf of Mexico beachesMar. Pollut. Bull.1001201532733310.1016/j.marpolbul.2015.08.029
- M.F.SimõesA.AntunesC.A.OttoniM.S.AminiI.AlamH.AlzubaidyV.B.BajicSoil and rhizosphere associated fungi in gray mangroves (Avicennia marina) from the Red Sea – a metagenomic approachGenomics Proteomics Bioinf.135201531032010.1016/j.gpb.2015.07.002
- C.Y.TurenneS.E.SancheD.J.HobanJ.A.KarlowskyA.M.KabaniRapid identification of fungi by using the ITS2 genetic region and an automated fluorescent capillary electrophoresis systemJ. Clin. Microbiol.376199918461851
- Y.WangW.P.ZhangH.L.CaoC.S.ShekR.M.TianY.H.WongP.-Y.N.A.QianDiversity and distribution of eukaryotic microbes in and around a brine pool adjacent to the Thuwal cold seeps in the Red SeaFront. Microbiol.5201410.3389/fmicb.2014.00037
- J.YangR.-H.JiaoL.-Y.YaoW.-B.HanY.-H.LuR.-X.TanControl of fungal morphology for improved production of a novel antimicrobial alkaloid by marine-derived fungus Curvularia sp. IFB-Z10 under submerged fermentationProcess Biochem.512201618519410.1016/j.procbio.2015.11.025
