Abstract
Eight isolates identified as belonging to the genus Bacillus were obtained from Aloqt (a crusty dried product made from goat milk). The cell-free culture supernatant (CFCS) from the eight isolates possessed an inhibitory spectrum against Staphylococcus aureus and Escherichia coli. The eight unknown bacterial isolates were identified by PCR amplification of their 16S ribosomal RNA gene and sequencing of the resulting PCR products. All of the isolates were classified as members of Bacillus amyloliquefaciens, as their 16S rDNA similarities to the respective species were greater than 99%. The phylogenetic analysis grouped two isolates with strain BCRC 11601 and the remaining six isolates with strains MPA 1034 and BCRC 11601. The inhibitory activity of the CFCS was either highly reduced or fully inactivated when treated by proteolytic enzymes, suggesting the possible involvement of a protein/polypeptide bacteriocin-like inhibitory substance (BLIS) in their antagonism. The optimum growth conditions for maximum inhibitory activity were achieved at an initial pH of 7, incubation temperature of 37 °C, and NaCl concentration of (1%). A considerable decrease and/or complete loss of activity occurred at values above and below the optimum. The maximum inhibitory activity occurred after 24 h of incubation; as the incubation time increased, a decrease in the activity was observed until a complete loss of activity occurred after 72 h of incubation.
1 Introduction
With the emergence of bacteria that are resistant to antibiotics, a marked increase has occurred in the level of attention given to bacteriocins [Citation1]. These proteins are considered ideal candidates for food preservation and personal care applications because the range of their activity is limited to closely related species, and therefore, they would theoretically have no harmful effects on humans and their normal microbiota [Citation2,Citation3].
Goat milk has a very rich and complex autochthonous microbiota [Citation4]. The autochthonous microbiota of raw goat milk is particularly interesting due to its diversity and the presence of several bacteriocin-producing bacteria [Citation5].
Bacteriocins are ribosomally synthesized proteins produced during the primary phase of growth by a diverse group of microorganisms that show bactericidal activity, usually against closely related species [Citation6]. The known biological mode of action of bacteriocins is related to their ability to bind rapidly to anionic liposomes [Citation7]. After bacteriocins interact with anionic lipids, they form pores in the lipid membrane by interacting with the peptidoglycan precursor lipid II, thus preventing peptidoglycan biosynthesis and disrupting the lipid bi-layer organization of the membrane [Citation8].
The majority of recent reports have focussed on the production by Gram-positive bacteria of antibacterial molecules categorized as bacteriocins [Citation9,Citation10], even though the majority of Gram-positive bacteria that produce bacteriocins, and have been described so far, are related to lactic acid bacteria [Citation11,Citation12]. However, its sensitivity to proteases, its low solubility above pH 6 and the emergence of resistant strains, indicate the need for an alternative to Gram-positive bacteria as the producer organisms [Citation13]. Bacillus represents an alternative genus to be investigated for the production of bacteriocins, including antimicrobial peptides with increased protease tolerance [Citation9,Citation10,Citation14]. Bacillus includes many industrial grade species, some of which have a history of being safely used in the food industry as probiotics [Citation15,Citation16].
Sequence-based genotyping and inferring phylogenetic relations among bacteria have been widely performed using 16S ribosomal RNA gene sequences [Citation16]. Bacterial 16S rRNA genes generally contain nine hypervariable regions flanked by conserved stretches, which enable PCR amplification when using universal primers for species identification [Citation17,Citation18].
The present study aimed to provide relevant information, including the proper identification and the phylogenic position of the antagonistic Bacillus spp. isolated from crusty dried goat milk. Additionally, the study aimed to verify the inhibitory spectrum, the nature of the produced compounds and the optimum growth conditions for the production of the isolated species.
2 Materials and methods
2.1 Sample collection and isolation
Eleven Aloqt (traditional crusty dried processed goat milk) samples were collected from different stores in the Jeddah area (approximately two samples per store), 10 g of each Aloqt sample were ground and homogenized together in 90 ml of sterile normal physiological saline solution (0.85% NaCl, w/v) and subjected to tenfold serial dilutions up to 10−9.
One ml of the selected dilutions (>10−4) from each sample was inoculated into two sets of sterile de Man, Rogosa and Sharpe (MRS) agar media (Difco™, Detroit, MI, USA). One set was incubated at 37 °C in 5% CO2 environment for 48 h, and the other set was incubated at the same temperature but under anaerobic conditions using GasPak (EZ™ Gas Generating Container Systems, BD – Becton, Dickinson and Co., Franklin Lakes, NJ, USA). After incubation, the samples were selected and subjected to Gram staining and examined under the microscope to allow visualization of the basic cell characteristics.
Bacterial colonies were further subjected to additional microbiological analysis for identification by observing their morphological, physiological and biochemical characteristics as described in previous reports [Citation19]. The biochemical characteristics were determined using both API 20E and API 50CH bacterial identification test strips (bioMerieux SA, France).
2.2 Screening of isolates for antibacterial activity
Isolates identified as belonging to the genus Bacillus (Gram-positive spore-forming rods) were screened using two different methods, cross-streak and agar-well diffusion inhibition assays [Citation20,Citation21], for their antibacterial activity against two pathogenic organisms (Staphylococcus aureus and Escherichia coli), which were obtained from the King Fahd Hospital in Al-Madina. Rapid screening for antagonistic isolates was carried out first by the cross-streak method. Briefly, Müller–Hinton agar plates (Oxoid Ltd, Basingstoke, Hampshire, UK) were prepared and inoculated with isolates of the genus Bacillus by a single streak in the center of the Petri dish and incubated at 37 °C for 48 h to provide the active organism with enough time to produce the antibiotic substance, which then diffuses into the agar medium. The plates were then seeded with indicator organisms by streaking perpendicular to the line of the Bacillus isolate growth. Antagonism was observed by the end of the incubation at 37 °C for 24 h based on the inhibitory interaction between the isolates and the indicator strains.
Based on the cross-streak method results, all isolates of the genus Bacillus showing antagonistic activity were cultured in sterilized Falcon tubes (Sigma–Aldrich, St. Louis, MO, USA) containing 10 ml of the MRS broth with the pH adjusted to ∼6.8 and incubated for 48 h at 37 °C in 5% CO2 environment (until approximately 106 CFU ml−1) to carry out the agar-well diffusion inhibition assay. After incubation, the bacterial cells were removed from the culture by centrifugation for 20 min at 4500 × g and 4 °C. Cell-free culture supernatant (CFCS) was harvested and filter-sterilized using 0.45 μm micro-filters (Acrodisc® 25 mm Syringe Filter w/0.2 μm Supor® Membrane, Pall Co., MI, USA) The obtained (CFCS) was further concentrated into 2-ml aliquots using a rotary evaporator (Rotavapor® R-200, BÜCHI Labortechnik, Flawil, Switzerland).
The indicator organisms (S. aureus and E. coli) were grown overnight on nutrient broth (Oxoid Ltd, Basingstoke, Hampshire, UK) according to their specific growth requirements. To create a seeded test media, the 18–24 h culture of the indicator strains was mixed with sterilized Müller–Hinton agar earlier cooled to 45 °C at a final concentration of ∼105 CFU ml−1, poured onto plates, allowed to solidify, and then dried in a sterile hood for approximately 2 h prior to use. Sterile Pasteur pipettes were used to punch 5-mm wells onto the surface of the Müller–Hinton agar plates seeded with the indicator organisms. Then, a micropipette (Eppendorf Biotech, Hamburg, Germany) was used to add 100 μl of each CFCS sample to the wells, where the samples remained at room temperature for 45–60 min to freely diffuse. All plates were then incubated overnight at the optimal growth temperature (37 °C) for the indicator organisms. The presence of inhibition halos (clear zones) around the well was recorded as the antimicrobial activity of the tested isolate. The well diffusion inhibition assays were all conducted in triplicates.
2.3 Molecular identification of the active antibacterial isolates
The identity of the bacterial isolates showing antibacterial activity was confirmed using molecular approaches. All isolates were cultured overnight in 10 ml Luria–Bertani (LB) broth (Difco™, Detroit, MI, USA) at 37 °C for 18 h. After incubation, the cellular DNA was extracted as previously described by Kageyama et al. [Citation22]. Briefly, 2-ml aliquots of the broth culture were collected in 1.5 ml micro-centrifuge tubes (Eppendorf Biotech, Hamburg, Germany) and centrifuged at 13,000 × g for 3 min. The pellet was then suspended in 200 μl TE buffer (10 mM Tris–HCl [pH 8] and 1 mM EDTA), 100 μl of lysozyme (20 mg/m), and 10 μl proteinase K (20 mg/ml), and the mixture was incubated for 60 min at 37 °C. Afterwards, 250 μl GPT reagent (6 M guanidine thiocyanate dissolved in 50 mM Tris/HCl, pH 8) and 5 μl RNase were added and incubated for 20 min at 65 °C. After incubation, 450 μl of Tris-buffered phenol (pH 8) was immediately added, and the tubes containing the mixture were placed in a boiling water bath for 15 min; then, 250 μl chloroform/isoamyl alcohol (24:1, v/v) was added to remove the excess proteins and lipids. Afterwards, the mixture was centrifuged for 10 min at 13,000 × g. The upper aqueous phase (∼500 μl) was transferred to a fresh tube and was mixed with an equal amount of 100% ethanol and a 1/10 volume of 3 M sodium acetate before being maintained at 20 °C for 1 h. Samples were centrifuged at 13,000 × g for 15 min at 4 °C, and the supernatant was then discarded. Traces of GPT reagent were removed by the addition of 500 μl ice-cold 70% ethanol to the nucleic acid pellet, and samples were centrifuged at 13,000 × g again for 5 min at 4 °C. The nucleic acid pellet was dried under a vacuum for 20 min and finally re-suspended in 50 μl of sterile nuclease-free TE buffer. Based on the UV absorbance at specific wavelengths, the molar concentration and purity of the nucleic acid DNA were determined using a spectrophotometer (AE-450, ERMA Inc., Tokyo, Japan). A final concentration of 5 μg/ml and purity indicated by an A260/A280 ratio of 1.8–2.1 were considered acceptable. All reagents used in the extraction of the cellular DNA were obtained from (BDH Chemicals Ltd, Poole, UK or ACROS Organics, New Jersey, USA).
A nearly complete 16S rRNA gene sequence was determined for each isolated strain. The 16S rDNA was amplified by polymerase chain reaction (PCR) using the prokaryotic 16S rDNA universal primers 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) forward primer and 1542R (5′-ACAAAGGAGGTGATC-3′) reverse primer. The PCR was performed with an automated DNA thermal cycler (Perkin-Elmer 2720, Applied Biosystems Inc., USA). The amplification cycle profile was as follows: an initial denaturation step at 94 °C for 5 min; 35 cycles of denaturation at 94 °C for 60 s, primer annealing at 60 °C for 60 s, and primer extension at 72 °C for 120 s; and a final extension step at 72 °C for 10 min.
The amplified PCR products were separated on a 1.5% (w/v) agarose gel (Sigma–Aldrich Chemical Co., St. Louis, USA) in an electrophoresis cell (Wide Mini-Sub Cell GT, Bio-Rad, UK) containing 1× Tris borate EDTA (TBE) buffer (0.09 M Tris, 0.09 M boric acid, and 20 mM EDTA, pH 8) (Gibco® Life Technologies, NY, USA) at a constant voltage of 5 V/cm (∼80–100 V) for 45–100 min. After electrophoresis, the agarose gel was stained with 0.5 μg/ml ethidium bromide (Invitrogen Life Technologies, CA, USA) for 20 min. The DNA bands were visualized and photographed under UV transillumination in a gel documentation system (GelDoc 2000, Bio-Rad, UK). The sizes of the resulting DNA fragments were determined by comparison with 1 Kb Plus DNA Ladder standard molecular weight marker (Invitrogen Life Technologies, CA, USA). Prior to the DNA sequencing, all PCR products were purified with Silica Spin Column (Pure-Link PCR Purification Kit, Invitrogen™, NY, USA). The PCR products were sequenced by Macrogen, Inc. (Seoul, Korea). A sequence database search using the BLAST search program analysis of the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/BLAST/) against various sequences was used to identify the phylogenetic similarities among the microbially active isolates and the published DNA sequences in GenBank.
2.4 Phylogenetic analysis
Sequence data from the microbially active isolates were used to construct a phylogenetic tree using the maximum likelihood method [Citation23]. The statistical significance of the tree topology was evaluated using a bootstrap analysis of the sequence data with clustalW software [Citation24]. The percentages of the replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches [Citation25]. Nucleotide substitution rates (Knuc values) were calculated, and the tree was drawn to scale, with branch lengths shown in the same units as those of the evolutionary distances used to infer the phylogenetic tree [Citation26]. The evolutionary history was inferred using the maximum likelihood method based on the Jukes–Cantor model [Citation27]. The tree with the highest log likelihood (−2076.9473) is shown. Initial tree(s) for the heuristic search were obtained automatically by applying the maximum parsimony method. A discrete gamma distribution was used to model evolutionary rate differences among sites (five categories (+G, parameter = 0.2891)). The analysis involved 22 nucleotide sequences. All positions containing gaps and missing data were eliminated. A total of 778 positions were included in the final dataset. Evolutionary analyses were conducted using the MEGA6 program [Citation28].
2.5 Enzymatic degradation to confirm the nature of the antimicrobial compound
The effect of different enzymes (all from Sigma–Aldrich, Inc., St. Louis, MO, USA) on the antimicrobial activity was determined [Citation29]. Briefly, aliquots (100 μl) of the CFCS were combined with various concentrations of the tested enzymes (0.1–5 mg/ml), and the mixtures were incubated overnight, with the pH and temperature adjusted to those optimal for the enzymatic activity. After 24 h, all samples were used to repeat another well diffusion inhibition assay against the indicator organisms.
2.6 Incubation time and antibacterial substance production
To decide the optimal incubation time for the maximum production of the antibacterial compound, a single representative isolate was selected, inoculated in MRS broth medium, and incubated for different time intervals of 12, 24, 48 and 72 h. After incubation, the cell density was measured at 600 nm using un-inoculated MRS broth as the blank. Then, the CFCS was harvested from all batches and used to conduct another well diffusion inhibition assay against the indicator organisms as previously explained.
2.7 Growth conditions for antibacterial substance production
To determine the optimum pH for the antibacterial substances production, three sets of 100 ml of MRS broth were prepared and adjusted to pH 4, 7 and 9, respectively with 6 M HCl or 6 M NaOH stock solutions. The flasks were inoculated with 18- to 24-hour-old Bacillus amyloliquefaciens culture and incubated at 37 °C for 24 h. After incubation, the CFCS was harvested from all batches, and the pH of the supernatants was readjusted to ∼6.8 with sterile NaOH to attain the maximum antibacterial activity before being used against the indicator organisms.
To optimize the effect of incubation temperature, flasks containing 100 ml of sterilized MRS broth with 18- to 24-hour-old B. amyloliquefaciens culture were incubated at 25, 37 and 45 °C for 24 h. After incubation, the CFCS was harvested and used against the indicator organisms.
Sodium chloride (NaCl) salt concentration was optimized by preparing four sets of 100 ml of sterile MRS broth, with salt concentrations of 1, 2, 4 and 6%, respectively. The flasks were inoculated and incubated at 37 °C for 24 h, as previously explained. After incubation, the CFCS was harvested and used in well diffusion inhibition assays.
3 Results and discussion
3.1 Characterization of the unknown isolates
A total of 54 bacterial isolates were successfully purified from the Aloqt using MRS agar plates. Sixty-two biochemical and physiological tests (API 50CH and 20E test strips) and a number of morphological characteristics (primarily colony, cell and spore morphology) were selected and investigated to provide effective differentiation of all isolates. Based on the results of the tests, all isolates were identified as belonging to the genus Bacillus. Although Aloqt (a type of processed milk) was purported to contain Lactobacillus spp., as observed in other studies [Citation4,Citation5], Bacillus was the only organism recovered from four individual lots of the product in this study. Similar results showing the isolation of the genus Bacillus from fermented milk products presumed to contain Lactobacillus spp. was observed in a previous report [Citation30]. Possibly, Lactobacillus isolates are not competitive with the strains present in the milk products. The slow growth of Lactobacillus strains, along with the presence of several microorganisms releasing metabolic products, may interfere with the metabolism of the Lactobacillus, thereby leading to the suppressive effects [Citation31]. Another explanation is that Lactobacillus strains often have poor stability during storage; in addition, the inclusion of flavors, such as essential oils, may affect Lactobacillus bacteria-containing dairy products because of the antagonistic action of the added flavors against several microorganisms [Citation31,Citation32].
Optimal growth conditions for the isolates were observed at temperatures between 35 °C and 37 °C, at pH 7, and under aerobic conditions. No growth occurred below 15 °C or above 50 °C, and no growth occurred under anaerobic conditions. On solid culture media, the colonies tended to spread quickly into lawn formation, with an extremely wrinkled texture and a thick, opaque slime that was secreted by the colonies. Gram staining and microscopic examination of each sample of bacterial growth revealed a Gram-positive, endospore-producing bacillus. Cells often form chains, and the spores appeared oval in shape and central, paracentral or sub-terminal in un-swollen sporangia.
3.2 Antimicrobial activity assay
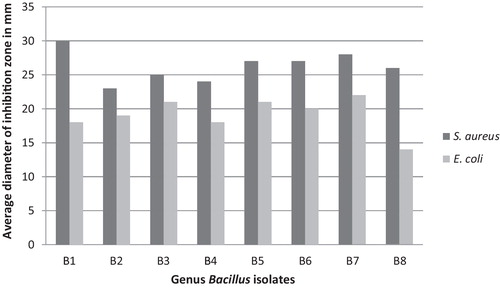
Eight of the 54 Bacillus isolates obtained from the Aloqt possessed potent antibacterial activity in the cross-streak assay. The well diffusion inhibitory assay revealed that the antibacterial compounds produced by the eight isolates exhibited various degrees of inhibitory activities against both the Gram-positive and Gram-negative pathogenic bacteria, S. aureus and E. coli, respectively, indicating that the isolates may have a broad spectrum of inhibition (). The assay also revealed that all isolates showed higher antibacterial activity towards S. aureus than E. coli, suggesting that the antibacterial substance produced by the isolates recovered in this study may be a type of bacteriocin-like inhibitory substance (BLIS), as bacteriocins produced by Gram-positive bacteria demonstrate high bactericidal activity directed principally against other Gram-positive bacteria, whereas Gram-negative bacteria are less sensitive to the bacteriocins produced by Gram-positive bacteria [Citation33].
3.3 Molecular identification of the isolates
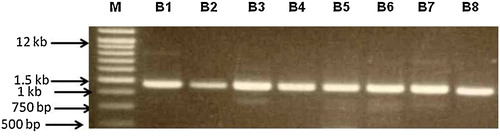
All eight isolates that presented antibacterial activity against the indicator strains were subjected to molecular identification using PCR amplification of almost the complete 16S rRNA gene. A single gene fragment was amplified from each isolate. The size of the generated fragments was in the range of 1.4–1.5 kb (). PCR was followed by DNA sequence analysis of the resulting PCR product. The 16S rDNA nucleotide sequences were determined for all eight isolates, and a database search was conducted. Sequencing of the 16S rRNA gene was sufficient to provide a proper and reliable identification of the isolates, with variations that allowed differentiation of the species. The BLAST search demonstrated that these isolates were closely related to B. amyloliquefaciens, with sequence similarity of more than 99% to the 16S rRNA gene of B. amyloliquefaciens, and this was sufficient to indicate that these eight isolates belong to the same species. Bacillus amyloliquefaciens is closely related to Bacillus subtilis and Bacillus licheniformis, and separation of these organisms solely on the basis of classical tests is not possible [Citation34]. The B. amyloliquefaciens isolates in our study were easily differentiated by using the 16S rDNA-based molecular identification technique.
Fig. 2 PCR amplification of the 16S rRNA gene from the eight microbially active isolates showing a single gene fragment in the range of 1.5 kb. Lane M: standard DNA molecular weight marker (1 kb plus DNA ladder), Lanes B1 to B8, the eight isolates.
Few reports exist on bacteriocin synthesis in B. amyloliquefaciens [Citation35]. Recently, B. amyloliquefaciens GA1 strains were identified as producers of a novel bacteriocin named amylolysin, which has potent antimicrobial activity towards the food-borne pathogen Listeria monocytogenes [Citation36] and has been found to be less sensitive to proteases than other bacteriocins produced by lactic acid bacteria [Citation37]. Interestingly, previous reports addressing the antibacterial spectrum of bacteriocins isolated from B. amyloliquefaciens showed antibacterial activity against closely related Gram-positive bacteria only, whereas no inhibition activity was observed against Gram-negative bacteria, especially E. coli [Citation37,Citation38]. However, the bacteriocin-like substance from B. amyloliquefaciens reported in this study showed activity against both Gram-positive and Gram-negative E. coli.
3.4 Phylogenetic affiliation of the isolates
The constructed phylogenetic tree using the 16S rRNA gene sequences of the related members from Bacillus (B. subtilis, B. licheniformis, B. tequilensis, B. cereus, B. anthracis, B. thuringiensis, B. methylotrophicus, B. megaterium and B. amyloliquefaciens) revealed that all eight isolates were grouped with B. amyloliquefaciens and are phylogenetically separated from other Bacillus species (). The phylogenetic tree clearly shows that the eight isolates could be divided into two closely related clades, separated by a short branch length, which indicates a limited number of nucleotide changes. Two isolates (B3 and B8) comprised one clade that was closely related to the B. amyloliquefaciens strain BCRC 11601 (GenBank accession NR116022.1). The remaining six isolates (B1, B2, B4, B5, B6, and B7) made another clade that was more closely related to strain MPA 1034 (GenBank accession NR117946). These results were supported by the BLAST search analysis against published 16S rRNA gene sequences in the GenBank database ().
Fig. 3 Likelihood phylogenetic tree based on the nucleotide sequences of the 16S rRNA gene, depicting the phylogenetic relationship between the eight isolates and other representatives of the Bacillus genera. All sequences were aligned on Streptomyces griseus (GenBank accession D63872.1). The tree was drawn to scale, with branch lengths measured in the number of substitutions per site. Bootstrap values from 500 replicates.
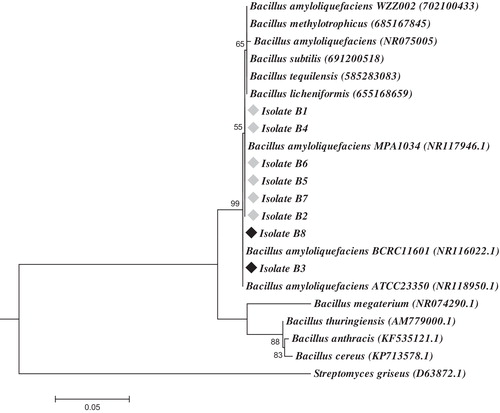
Table 1 BLAST search results of the studied isolates with the first species on the NCBI including their sequences accession numbers and similarities.
To define the borders of the variable region(s) among the two divided clades, multiple sequence alignment was performed using the 16S rRNA gene sequences of the eight studied isolates, along with B. amyloliquefaciens strains MPA 1034 and BCRC 11601. The alignment results revealed that 16S rRNA gene sequences were 100% identical among isolate B3; isolate B8 with strain BCRC 11601; and isolates B1, B2, B4, B5, B6, B7 with strain MPA 1034. The alignment results defined the variation between the two divided clades as a single nucleotide polymorphism (SNP) at position 107 (Supplemented Fig. S1). This intra-genomic heterogeneity in the 16S rRNA gene among the different strains of the Bacillus spp. has been previously reported [Citation39].
3.5 Effect of enzyme digestion on the antibacterial activity
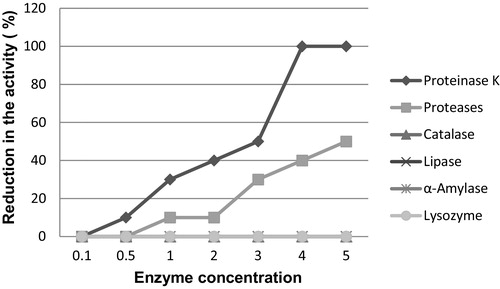
The inhibitory nature of bacterial supernatants may result from hydrogen peroxide elaborated by the bacteria [Citation20]. To rule out hydrogen peroxide inhibition culture, the supernatant was treated with catalase enzyme. After treatment of the test culture supernatant with catalase, no change in inhibitory activity of the isolates was observed, indicating that the active agent is not hydrogen peroxide being elaborated by the individual isolates.
The nature of the antibacterial substance was confirmed by enzymatic assays on the cell-free culture of the antagonistic isolates (). The antibacterial activity of the cell-free culture supernatant was not affected by treatment with lipase, α-amylase or lysozyme, which suggested that the activity of the compound is not dependent on glycosylation and that no carbohydrates and lipids are present in its structure. However, the activity was completely lost after the treatment with proteinase K, and similar results have been reported by many investigators regarding the characterization of the bacteriocins [Citation40]. Hence, possibly, the peptide/protein nature of the molecules is involved in the inhibition. The results reported herein can be considered as indirect evidence that the antagonism displayed by the B. amyloliquefaciens bacterial isolates is mediated by bacteriocin or bacteriocin-like inhibitory substances (BLIS).
In addition, bacteriocins usually have low molecular weight (rarely over 10 kDa) and can be easily degraded by proteolytic enzymes, especially by the proteases of the mammalian gastrointestinal tract [Citation2,Citation41]. This understanding is in agreement with the observed results in this study because the activity of the antibacterial compound produced by the tested isolates was significantly decreased after treatment with other protease enzymes (pepsin, trypsin and chymotrypsin); thus, these antibacterial compounds would not interfere with the intestinal microbiota and would be safe for human consumption.
3.6 Effect of incubation time
Representative isolate B1 was selected to test the effect of incubation time on the antibacterial activity. The maximum antibacterial compound production occurred during the early stationary phase (24 h). During the extended stationary phase of incubation, the activity of the compound decreased considerably to a complete cessation of activity (72 h). Although an increase was observed in the number of cells, no activity was observed against the indicator isolate after 72 h of incubation ().
Fig. 5 Effect of incubation time on inhibitory activity of the antibacterial compound shown against Staphylococcus aureus, with the increase in bacterial cell number measured at OD 600 nm.
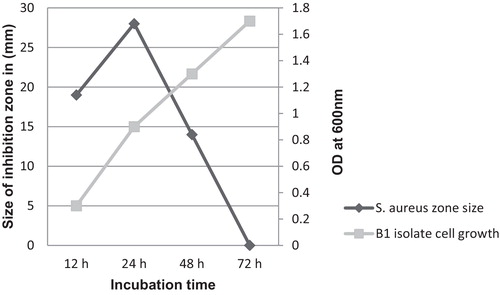
This decrease in activity, as described by previous reports, may result from the increase in the concentration of the BLIS over time, leading to protein aggregation and adsorption to the cell surface, followed by a feedback regulation that inhibits the release of more BLIS [Citation42]. Furthermore, because B. amyloliquefaciens is responsible for much of the protease enzymes production [Citation43], proteolytic degradation of the remaining BLIS in the culture media will take place by the action of the endogenous extracellular proteases induced during this growth phase, thereby resulting in the complete loss of activity.
3.7 Optimum growth conditions for antibacterial activity
Bacteriocins of Gram-positive bacteria have a concentration-dependent mode of action, which is affected by the various physiological factors that affect cell growth, such as pH, temperature and salt concentration; all of these factors seem to affect bacteriocin production, as well as its activity [Citation44].
The highest antibacterial activity for all isolates against indicator strains was recorded in MRS broth with an adjusted pH of 7, a salt concentration of 1%, and an incubation temperature of 37 °C (). A considerable decrease was observed in the antibacterial activity of the isolates, but the antibacterial activity remained relatively stable against both indicator strains after incubation at pH 11. These findings are in agreement with previous reports of considerable inactivation of bacteriocin action at pH values above 9 [Citation45]. The results also show that the antibacterial activity of the isolates survived incubation at acidic (pH 4) pH levels, suggesting that the producing isolates have some acid-tolerance mechanism that supports their BLIS production even under extremely acidic conditions.
Fig. 6 The effect of different physiological factors on the antibacterial activity of Bacillus amyloliquefaciens cell-free culture supernatant tested against Staphylococcus aureus (panels A, C and E) and Escherichia coli (panels B, D and F).
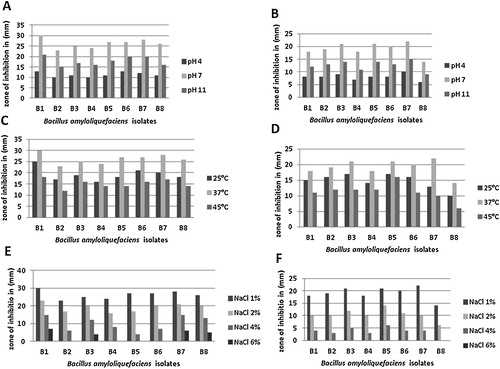
The optimum temperature for the production of the antibacterial compound was 37 °C, indicating that the optimum temperatures for production and growth are correlated. A significant reduction in the antibacterial activity among all isolates against the indicator strains was observed on incubation at 45 °C; by contrast, the reduction in the antibacterial activity observed on incubation at 25 °C was minimal (). Similar findings were also recorded in previous research [Citation46].
The data indicate that the antibacterial activity of the isolates survived salt concentrations up to 4%, which allows the hypothesis that salt-tolerance mechanisms exist in the produced isolates, which helps maintain the osmolarity of the cell.
The antibacterial activity was observed against S. aureus at all of the tested NaCl concentrations (1–6%, w/v), but in the case of E. coli, a complete loss of antibacterial activity against Escherichia coli occurred when the 6% (w/v) NaCl concentration was used (). Probably, the bacterial metabolism is sensitive to salt, as salt exhibits specific ionic and water binding properties [Citation47]; thus, the addition of salt leads to a decrease in the water activity (aw). A decrease in aw below the optimum values for growth often result in a linear decrease of the growth rate and, thus, in the production of the antibacterial compound [Citation48,Citation49].
Although, all of the antagonistic isolates in this study belonged to the same species, the antibacterial compound produced by them showed a wide range of activity against the indicator strains ( and ). This result was in agreement with a previous report regarding BLIS produced by B. amyloliquefaciens isolates [Citation50], and based on the bacteriocin concentration-dependent mode of action theory, the difference in the susceptibility patterns of the test strains could be due to differences in the amounts of inhibitory substance produced among the isolates.
Overall, the results definitely suggest that the active component of the producer isolates could be an antibacterial molecule of bacteriocin-like inhibitory substances. Such data can justify further studies with the purified compound to confirm their role as a bacteriocin, as well as their potential to be employed in different applications.
4 Conclusion
According to the results collected during the above-described survey, the antibacterial activity of the isolates was properly characterized and verified. These antagonistic isolates possessed an inhibitory effect against both Gram-positive and Gram-negative indicator organisms (S. aureus and E. coli, respectively) and thus may represent a potential source for the production of a wide inhibitory spectrum antibiotic. The molecular approaches used in this study were successfully able to identify all of the isolates as B. amyloliquefaciens and reveal the intra-genomic heterogeneity in their 16S ribosomal RNA genes, which classified them as two different strains: BCRC 11601 and MPA 1034. Finally, the nature of the produced compound, along with the optimum growth conditions for its maximum production, was established, and indirectly, the compound has been proven to be a type of bacteriocin or bacteriocin-like inhibitory substance (BLIS). These findings open several perspectives for further investigation and prompt new interest in antimicrobial substances produced by B. amyloliquefaciens bacteria.
Supplementary data
Download PDF (1.4 MB)Acknowledgments
This study was supported by a grant from the Ministry of Education and the Deanship of Scientific Research, Saudi Arabian government, which was provided to the Department of Biology, Faculty of Science, King Abdulaziz University. In addition the authors would like to thank Dr. Wael S. El-Sayed, Department of Biology, Faculty of Science, Taibah University, for providing us with the prokaryotic 16S rRNA gene universal primers.
Notes
Peer review under responsibility of Taibah University.
References
- A.GravesenK.SørensenF.M.AarestrupS.KnøchelSpontaneous nisin-resistant Listeria monocytogenes mutants with increased expression of a putative penicillin-binding protein and their sensitivity to various antibioticsMicrob. Drug Resist.72001127135
- L.H.DeeganP.D.CotterC.HillP.RossBacteriocins: biological tools for bio-preservation and shelf-life extensionInt. Dairy J.16200610581071
- C.E.ShelburneF.Y.AnV.DholpeA.RamamoorthyD.E.LopatinM.S.LantzThe spectrum of antimicrobial activity of the bacteriocin subtilosin AJ. Antimicrob. Chemother.592007297300
- L.PsoniC.KotzamanidisM.YiangouN.TzanetakisE.Litopoulou-TzanetakiGenotypic and phenotypic diversity of Lactococcus lactis isolates from Batzos, a Greek PDO raw goat milk cheeseInt. J. Food Microbiol.1142007211220
- S.SchirruS.D.TodorovL.FavaroN.P.MangiaM.BasagliaS.CasellaR.ComunianB.FrancoP.DeianaSardinian goat's milk as source of bacteriocinogenic potential protective culturesFood Control252012309320
- D.DriderG.FimlandY.HéchardL.M.McMullenH.PrévostThe continuing story of class IIA bacteriocinsMicrobiol. Mol. Biol. Rev.702006564582
- H.BrötzG.BierbaumA.MarkusE.MolitorH.G.SahlMode of action of the lantibiotic mersacidin: inhibition of peptidoglycan biosynthesis via a novel mechanismAntimicrob. Agents Chemother.391995714719
- G.PattonK.A.DonNew developments in lantibiotic biosynthesis and mode of actionCurr. Opin. Microbiol.82005543551
- J.WangL.ZhangK.TengS.SunZ.SunJ.ZhongCerecidins a novel lantibiotics from Bacillus cereus with potent antimicrobial activityAppl. Environ. Microbiol.80201426332643
- N.KhochamitS.SiripornadulsilP.SukonW.SiripornadulsilAntibacterial activity and genotypic–phenotypic characteristics of bacteriocin-producing Bacillus subtilis KKU213: potential as a probiotic strainMicrobiol. Res.17020153650
- E.RodriguezM.I.MartinezN.HornH.M.DoddHeterologous production of bacteriocins by lactic acid bacteriaInt. J. Food Microbiol.802003101116
- L.SaavedraP.CastellanoF.SesmaPurification of bacteriocins produced by lactic acid bacteriaMethods Mol. Biol.2682004331336
- M.BonnetM.M.RafiM.L.ChikindasT.J.MontvilleBioenergetic mechanism for nisin resistance, induced by the acid tolerance response of Listeria monocytogenesAppl. Environ. Microbiol.72200625562663
- J.DischingerM.JostenC.SzekatH.G.SahlG.BierbaumProduction of the novel two-peptide lantibiotic lichenicidin by Bacillus licheniformis DSM 13PLoS ONE42009e6788
- S.M.CuttingBacillus probioticsFood Microbiol.282011214220
- J.E.ClarridgeIIIImpact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseasesClin. Microbiol. Rev.172004840862
- K.M.McCabeY.H.ZhangB.L.HuangE.A.WagarE.R.McCabeBacterial species identification after DNA amplification with a universal primer pairMol. Genet. Metab.661999205211
- G.C.BakerJ.J.SmithD.A.CowanReview and re-analysis of domain-specific 16S primersJ. Microbiol. Methods552003541555
- A.G.O’donellJ.R.NorrisR.C.BerkeleyD.ClausT.KanekoN.A.LoganR.NozakiCharacterization of Bacillus subtilis, Bacillus pumilus, Bacillus licheniformis, and Bacillus amyloliquefaciens by pyrolysis gas–liquid chromatography, deoxyribonucleic acid–deoxyribonucleic acid hybridization, biochemical tests, and API systemsInt. J. Syst. Bacteriol.301980448459
- M.B.HanlinN.KalchayanandP.RayB.RayBacteriocins of lactic acid bacteria in combination have greater antibacterial activityJ. Food Prot.561993252255
- L.M.CintasJ.M.RodriguezM.F.FernandezK.SlettenI.F.NesP.E.HernandezH.HoloIsolation and characterization of pediocin L50, a new bacteriocin from Pediococcus acidilactici with a broad inhibitory spectrumAppl. Environ. Microbiol.61199526432648
- A.KageyamaK.TorikoeM.IwamotoJ.MasuyamaY.ShibuyaH.OkazakiK.YazawaS.MinotaR.M.KroppenstedtY.MikamiNocardia arthritidis sp. nov., a new pathogen isolated from a patient with rheumatoid arthritis in JapanJ. Clin. Microbiol.42200423662371
- J.FelsensteinEvolutionary trees from DNA sequences: a maximum likelihood approachJ. Mol. Evol.171981368376
- J.D.ThompsonD.G.HigginsT.J.GibsonCLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choiceNucleic Acids Res.22199446734680
- J.FelsensteinConfidence limits on phylogenies: an approach using the bootstrapEvolution391985783791
- M.KimuraT.OhtaFixation time of over dominant alleles influenced by random fluctuation of selection intensityGenet. Res.20197217
- H.KishinoT.MiyataM.HasegawaMaximum likelihood inference of protein phylogeny and the origin of chloroplastsJ. Mol. Evol.301990151160
- K.TamuraG.StecherD.PetersonA.FilipskiS.KumarMEGA6: molecular evolutionary genetics analysis version 6.0Mol. Biol. Evol.30201327252729
- C.B.LewusS.SunT.J.MontvilleProduction of an amylase sensitive bacteriocin by an atypical Leuconostoc paramesenteroides strainAppl. Environ. Microbiol.581992143149
- K.E.SutyakR.E.WirawanA.A.AroutchevaM.L.ChikindasIsolation of the Bacillus subtilis antimicrobial peptide subtilosin from the dairy product-derived Bacillus amyloliquefaciensJ. Appl. Microbiol.104200810671074
- I.SodiniA.LucasM.N.OliveiraF.RemeufG.CorrieuEffect of milk base and starter culture on acidification, texture, and probiotic cell counts in fermented milk processingJ. Dairy Sci.85200224792488
- C.M.MoritzV.L.RallM.J.SaekiA.F.JúniorInhibitory effect of essential oils against Lactobacillus rhamnosus and starter culture in fermented milk during its shelf-life periodBraz. J. Microbiol.43201211471156
- N.KayalvizhiP.GunasekaranProduction and characterization of a low-molecular-weight bacteriocin from Bacillus licheniformis MKU3Lett. Appl. Microbiol.472008600607
- F.G.PriestM.GoodfellowL.A.ShutelR.C.BerkeleyBacillus amyloliquefaciens sp. nov. norn. rev.Int. J. Syst. Bacteriol.3719876971
- A.Arguelles-AriasM.OngenaB.HalimiY.LaraA.BransB.JorisP.FickersBacillus amyloliquefaciens as a source of potent antibiotics and other secondary metabolites for biocontrol of plant pathogensMicrob. Cell Fact.820096364
- B.HalimiC.DortuA.Arguelles-AriasP.ThonartB.JorisAntilisterial activity on poultry meat of amylolysin, a bacteriocin from Bacillus amyloliquefaciens GA1Probiotics Antimicrob. Proteins2010120125
- A.Arguelles-AriasM.OngenaB.DevreeseM.TerrakB.JorisP.FickersCharacterization of amylolysin, a novel lantibiotic from Bacillus amyloliquefaciens GA1PLoS ONE82013e83037
- R.ScholzJ.VaterA.BudiharjoZ.WangY.HeK.DietelT.SchweckeS.HerfortP.LaschR.BorrissAmylocyclicin, a novel circular bacteriocin produced by Bacillus amyloliquefaciens FZB42J. Bacteriol.196201418421852
- T.CoenyeP.VandammeIntragenomic heterogeneity between multiple 16S ribosomal RNA operons in sequenced bacterial genomesFEMS Microbiol. Lett.22820034549
- M.ZamfirR.CallewaertP.C.CormeaL.De-VuystPurification and characterization of a bacteriocin produced by Lactobacillus acidophilus IBB 801J. Appl. Microbiol.872000923931
- R.BrombergI.MorenoR.R.DelboniH.C.CintraP.T.OliveiraCharacteristics of the bacteriocin produced by Lactococcus lactis subsp. cremoris CTC 204 and the effect of this compound on the mesophilic bacteria associated with raw beefWorld J. Microbiol. Biotechnol.212005351358
- L.De VuystR.CallewaertK.CrabbePrimary metabolite kinetics of bacteriocin biosynthesis by Lactobacillus amylovorus and evidence for stimulation of bacteriocin production under unfavorable growth conditionsJ. Microbiol.1421996817827
- S.J.ChoS.H.OhR.D.PridmoreM.A.JuilleratC.H.LeePurification and characterization of proteases from Bacillus amyloliquefaciens isolated from traditional soybean fermentation starterJ. Agric. Food Chem.51200376647670
- I.M.AasenT.MoretoT.KatlaL.AxelssonI.StorroInfluence of complex nutrients, temperature and pH on bacteriocin production by Lactobacillus sakei CCUG 42687Appl. Microbiol. Biotechnol.532000159166
- A.GalvezH.AbriouelR.L.LopezBacteriocin-based strategies for food bio-preservationInt. J. Food Microbiol.12020075170
- S.D.TodorovL.M.DicksInfluence of growth conditions on the production of a bacteriocin by Lactococcus lactis subp. lactis ST 34BR, a strain isolated from barley beerJ. Basic Microbiol.442004305316
- H.KorkealaT.T.AlankoT.TiusanenEffect of sodium nitrite and sodium chloride on growth of lactic acid bacteriaActa Vet. Scand.3319922732
- T.A.McMeekinR.E.ChandlerP.E.DoeC.D.GarlandJ.OlleyS.PutroD.A.RatkowskyModel for combined effect of temperature and salt concentration/water activity on the growth rate of Staphylococcus xylosusJ. Appl. Bacteriol.621987543550
- P.UguenJ.HamelinJ.P.Le-PennecC.BlancoInfluence of osmolarity and the presence of an osmoprotectant on Lactococcus lactis growth and bacteriocin productionAppl. Environ. Microbiol.651999291293
- L.BrittesK.CaumoA.BrandelliM.BrittesBacteriocin-like substance from Bacillus amyloliquefaciens shows remarkable inhibition of Acanthamoeba polyphagaParasitol. Res.1082011687691
Appendix A
Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jtusci.2016.02.007.
Appendix A
Supplementary data
The following are the supplementary data to this article: