Abstract
The New Delhi metallo-β-lactamase (NDM-1) is one of the most important resistance traits in Enterobacteriaceae. We characterized nine blaNDM-1 producing Enterobacteriaceae recovered from seven patients who have recently travelled or been treated in India (n=1) or mainland China (n=6) during December 2010–May 2012. All the China-linked patients had no links to the Indian subcontinent. The blaNDM-1 carrying plasmids belonged to the novel IncX3 (∼50 kb, in seven isolates including two Escherichia coli, two Klebsiella pneumoniae, one Citrobacter freundii, one Enterobacter aerogenes and one E. cloacae), IncA/C2 (∼140 kb, in one E. coli) or FII-F1B groups (∼110 kb, in one E. coli). Restriction fragment length polymorphism analysis of the seven IncX3 plasmids revealed identical pattern in six and two bands difference in the remaining one. The IncX3 plasmids carrying blaNDM-1 were epidemiologically linked to Guangzhou (n=1), Hunan (n=4), Haifeng (n=1) and Dongguan (n=1) in mainland China. Complete sequencing of the IncX3 plasmid pNDM-HN380 revealed that it was 54 035 bp long and encoded 52 open reading frames. The blaNDM-1 gene was found in a transposon-like structure flanked by ISAba125 and IS26, inserted into the plasmid genetic load region. The sequences of the blaNDM-1 containing module within the two IS elements were identical to those previously described for blaNDM-1-positive Tn125 in the plasmids or chromosome of Acinetobacter isolates. In summary, this is the first description of IncX3 plasmids carrying blaNDM-1. The findings indicate the worrisome involvement of an epidemic plasmid in the dissemination of NDM-1 in China.
Introduction
Carbapenem hydrolyzing β-lactamases are a major health threat in the management of gram-negative infections. In 2008, a novel type of carbapenemases, termed New Delhi metallo-β-lactamases (NDM-1) was identified in Escherichia coli and Klebsiella pneumoniae isolated in Sweden from a patient transferred 1 day previously from India.Citation1 In 2010, a landmark study identified 37 NDM-1 isolates in the UK, and 143 isolates in different parts of India, Pakistan and Bangladesh and demonstrated an epidemiological link to the Indian subcontinent.Citation2 In an environmental study conducted in 2010, NDM-1 producing bacteria of multiple species were grown from 12 of 171 seepage samples and 2 of 50 water samples collected in New Delhi.Citation3 Taken together, the available findings suggest that the Indian subcontinent is an important reservoir for NDM-1. Recently, small numbers of NDM-1-producing Enterobacteriaceae or Acinetobacter isolates have been identified in the Balkan states (Bosina, Kosovo, Montenegro and Serbia),Citation4,Citation5,Citation6,Citation7 the Middle EastCitation8,Citation9 and ChinaCitation10,Citation11,Citation12 among patients without obvious links to the Indian subcontinent.
The progenitor of blaNDM-1 remains undefined although similarity with the β-lactamase II from Erythrobacter litoralis has lead to proposal for an environmental reservoir, but this is disputed by others.Citation13,Citation14 Organisms that naturally produce carbapenems and plant pathogens are additional possibilities, but further work is required for confirmation.Citation15,Citation16 blaNDM-1 has always been found in association with an upstream insertion sequence ISAba125 which provides the −35 promoter sequence.Citation17 The dissemination of NDM-1 mainly involves mobile genetic elements rather than clonal spread. In Enterobacteriaceae, blaNDM-1 has been identified on plasmids with a narrow (IncF1B, IncFII) or broad (IncA/C, IncH, IncL/M and IncN) host range and rarely in the chromosome.Citation18,Citation19,Citation20,Citation21 The first plasmid to be completely sequenced was pNDM-HK (IncL/M, INSDC-GenBank accession HQ451074). The other plasmids that have been completely sequenced and deposited in the INSDC-GenBank were pNDM-1_Dok01 (IncA/C2, AP012208), pNDM-KN (IncA/C2, JN157804), pNDM10505 (IncA/C2, JF503991), pNDM10469 (IncA/C2, JN861072), pNDM102337 (IncA/C2, JF714412), pMR0211 (IncA/C2, JN687470), p271A (IncN2, JF785549), pNDM-MAR (IncH1B-F1B, JN420336) and pGUE-NDM (IncFII, JQ364967). Complete sequencing of plasmids provides information for the analysis of the genetic environment of blaNDM-1 and for a better understanding of the epidemiological aspects of plasmids. Previous studies have indicated that the blaNDM-1 gene was sometimes carried by untypeable plasmids.Citation2,Citation11 In this study, we characterized untypeable plasmids carrying blaNDM-1 in Enterobacteriaceae strains recovered from patients with an epidemiological link to mainland China. The results indicate the emergence of a novel plasmid carrying blaNDM-1 in multiple provinces in China.
Materials and methods
Bacterial strains, identification and antimicrobial susceptibility testing
The isolates included in this study were identified through a national program introduced since December 2010 for surveillance of carbapenem-resistant Enterobacteriaceae in Hong Kong, China. In short, admission screening was implemented for all inpatients with a recent history of hospitalization or surgery abroad. Fecal samples or rectal swabs were collected at admission and were plated onto MacConkey plates supplemented with 1 µg/ml meropenem (MCA-M). Colonies on the MCA-M were identified to species level. The combined disc method was used to screen for possible carbapenemase production by testing with ertapenem, imipenem and meropenem alone and in combination with ethylenediaminetetraacetic acid (Sigma, St Louis, MO, USA) or phenylboronic acid (Sigma).Citation22 An increase of ≥5 mm in presence of ethylenediaminetetraacetic acid or phenylboronic acid was used to indicate the possible presence of metallo-β-lactamase and class A carbapenemase, respectively. Isolates tested positive in the phenotypic assays were referred to a centralized laboratory for carbapenemase genes detection including blaNDM. During December 2011–May 2012, the program identified nine blaNDM positive Enterobacteriaceae isolates from seven patients. The nine isolates were included in the present study. Four of the isolates were recovered from two members of the same family.Citation11
The VITEK GNI system (bioMerieux Vitek Inc., Hazelwood, MO, USA) was used for bacterial identification. Susceptibility testing of the isolates was performed by disk diffusion assay and E-test (AB Biodisk, Solna, Sweden) and result interpreted according to the Clinical and Laboratory Standards Institute.Citation23
Carbapenemase gene detection
The major carbapenemase gene (blaNDM, blaIMP blaVIM, blaKPC and blaOXA-48) were detected by PCR using previously described primers.Citation11,Citation24,Citation25,Citation26 The entire coding sequence of blaNDM was amplified and sequenced using the following primer pairs: NDM-FW-10319 5′-GCC ATG TCA CTG AAT ACT CGT-3′ and NDM-RV-11450, 5′-GCG ATC CTT CCA ACT CGT-3′.
Multilocus sequence typing
The sequence type of K. pneumoniae and E. coli isolates was determined using the Pasteur Institute and University College Cork scheme, respectively.Citation27,Citation28
Plasmid studies
The transferability of blaNDM was tested by filter mating E. coli J53 Azr as the recipient. Transconjugants were selected on MacConkey medium containing sodium azide (100 µg/ml) and meropenem (0.5 µg/ml). In each set of experiment, absence of growth of the parent and the recipients in the selective agar plate was confirmed. Plasmid DNA was extracted with QIAGEN Midi Kit (Qiagen, Hilden, Germany) and introduced to competent E. coli DH5α (Invitrogen, Carlsbad, CA, USA) by electroporation, followed by selection of transformants on Luria Bertani agar supplemented with meropenem (0.5 µg/ml). The size of plasmids in the transconjugants or transformants was sized by S1-PFGE.
Replicon typing was conducted on transconjugant or transformant with a single plasmid encoding blaNDM. The polymerase chain reaction (PCR)-based replicon typing scheme was used for recognition of the following plasmid incompatibility groups (Inc): FIA, FIB, FIC, FIIA, HI1, HI2, I1-Iγ, L/M, N, P, W, T, A/C, K, B/O, X, Y and F.Citation29 The IncF plasmids were subtyped by sequencing.Citation30 The revised IncX plasmid replicon typing procedure was used for detection of the IncX1–IncX4 subtypes.Citation31 In all the isolates, the replicon location in the plasmids was confirmed by hybridization with probes specific for blaNDM and rep amplified by PCR from different samples.
The plasmids carrying blaNDM were further analyzed by restriction fragment length polymorphism (RFLP). Purified plasmid DNA was separately digested with EcoR1 and PstI (Takara, Dalian, China) in accordance with the manufacturer’s recommendation.
Plasmid sequencing
The complete sequence of the plasmid pNDM-HN380 carrying blaNDM in a DH5α transformant (originating from K. pneumoiae strain CRE380) was obtained by using the 454 GS FLX system (Roche, Branford, CT, USA) according to the manufacturer’s instruction. Plasmid DNA was prepared as previously described.Citation26 The library yield a total of 70 651 reads with average read length of 500 bp. The reads were assembled by the GS de novo Assembler (version 2.6) into five contigs. The gaps were closed by PCR and Sanger sequencing (Supplementary Table S1). The complete plasmid sequence was confirmed by comparison of the in silico RFLP and the experimental RFLP using EcoR1 and PstI restriction enzymes. The plasmid was annotated by RAST Server and each predicted open reading frames (ORFs) was further blast against the National Center for Biotechnology Information non-redundant protein database using BLASTP.Citation32 The WebACT and Mauve (version 2.2.0) softwares were used for alignment and comparison of plasmid sequences.Citation26,Citation33 XplasMap (version 9.0) was used for construction of a schematic plasmid map.Citation31
Results
Patient demographics and strains characteristics
The patient history and characteristics of the bacterial strains were summarized in . All patients had travel history and all but one of them had recently been hospitalized in mainland China before the blaNDM-producing strains were detected in Hong Kong. One isolate (CRE727) was identified in a urine sample. All the other isolates were identified in rectal swab or stool samples. All strains exhibited resistance to cephalosporins (ceftriaxone, ceftazidime), carbapenems (ertapenem, imipenem, meropenem), β-lactam/β-lactamase inhibitors combinations (amoxicillin-clavulanate, piperacillin-tazobactam). Coresistance involving multiple non-β-lactam drugs was common. Combined disc testing revealed that all had a metallo-β-lactamase phenotype. In all the strains, PCR and sequencing confirmed presence of the blaNDM-1 allele (100% identity to INSDC-GenBank HQ451074). Plasmid replicon typing showed that the blaNDM-1-carrying plasmids (50-140 kb in sizes) were of IncFIIY/FIBS IncX3, or IncA/C. In seven strains originating from five patients with history of medical care in Guangdong (Guangzhou, Haifeng and Dongguan) and Hunan provinces of China, the blaNDM-1 genes were localized to IncX3 plasmids of the same size (50 kb). In conjugation experiments, the plasmids harbouring blaNDM-1 in all nine strains could be transferred at frequencies of 10−1 to 10−5 transconjugants per donor cells. Transfer of the IncFIIY/FIBS and IncA/C2 carrying blaNDM-1 was associated with co-resistance to gentamicin, amikacin and/or tetracyclines in the recipients. No coresistance to non-β-lactam agents was found in recipients of the IncX3 plasmids carrying blaNDM-1.
Table 1 Patient demographics, bacterial strains and features of plasmids carrying blaNDM-1
RFLP analysis of IncX3 plasmids
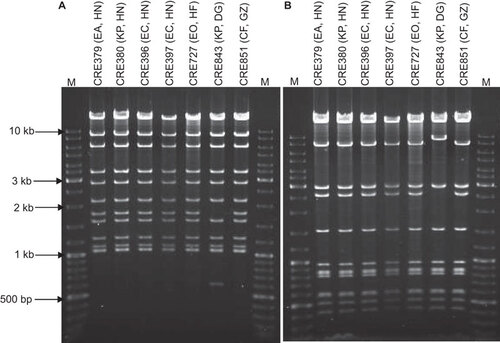
The IncX3 plasmids from the seven strains were subjected to RFLP analysis. Six plasmids had identical patterns after EcoR1 or Pst1 digestion (). The plasmid from K. pneumoniae strain CRE843 yield results that differed from that for the other strains by two bands for both restriction enzymes.
Figure 1 Restriction analysis of IncX3 plasmids carrying blaNDM-1. Plasmids were digested with (A) EcoRI and (B) PstI and separated by electrophoresis in 1% agarose. M, GeneRulerTM DNA ladder. The labels above each lane show the strain number, bacterial species origin (EA, E. aerogenes; KP, K. pneumoniae; EC, E. coli; EO, E. cloacae; CF, C. freundii) and the geographic source of importation (HN, Hunan; HF, Haifeng; DG, Dongguan; GZ, Guangzhou).
Sequence analysis of pNDM-HN380
The complete sequence of the plasmid, pNDM-HN380 originating from K. pneumoniae strain CRE380 was obtained (INSDC-GenBank accession JX104760). It is a 54 035 bp circular plasmid with an average GC content of 49% and 52 putative ORFs. showed a linear comparison with two other completely sequenced IncX3 plasmids (pEC13_35 and pIncX-SHV). The 30.2 kb backbone structure of pNDM-HN380 is typical of those described for IncX plasmid. The following set of core genes were shared among the three IncX3 plasmids: replication (replication initiation protein, pir; replication accessory protein, bis), partitioning (parA), plasmid maintenance (a putative DNA-binding protein, hns; a putative type III topoisomerase, topB), conjugation/type IV secretion system (T4SS, with 11 genes, pilX1 to pilX11), transcriptional activator (actX) and putative DNA transfer proteins (taxA and taxC). The 30.2 kb backbone of pNDM-HN380 is highly homologous to pIncX-SHV (100% coverage and 99% nucleotide identity); the similarity with that in pEC14_35 was lower (89% coverage and 98% identity).
Figure 2 Comparative analysis of (A) linear plasmid maps for three IncX3 plasmids, pEC14_35, pIncX-SHV, pNDM-HN380 and two blaNDM-1-carrying transposon sequences in pNDM-BJ01 and A. baumannii strain 161/07; (B) sequences downstream of insE and (C) sequences upstream of the ISAba125 in the 5′ end of blaNDM-1. The function blocks of the plasmids are indicated above the linear maps. The lengths of the ORFs are drawn in proportion to the size of the ORFs. Homologous ORFs in the plasmid maps are represented in the same colour. Direct repeats and mobile elements are labelled in blue and red, respectively. (B, C) Consensus regions in the aligned sequences of pNDM-HN380, pNDM-BJ01 and 161/07 are marked with asterisk. The sequences identical in pNDM-HN380 and pNDM-BJ01 are coloured green. The ORFs are indicated by grey shading and the arrow next to the label indicates the ORF orientation. The accession numbers were: pEC14_35 (JN935899), pIncX-SHV (JN247852), pNDM-HN380 (JX104760), pNDM-BJ01 (JQ001791) and Acinetobacter baumannii strain 161/07 (HQ857107).
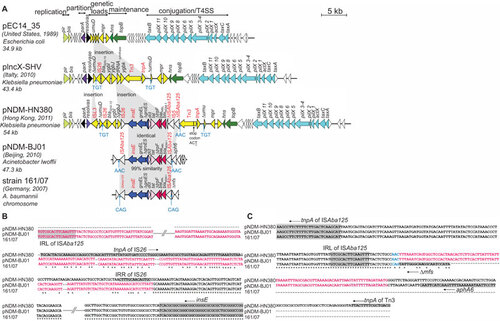
The genetic load region between the resolvase, res gene and the hns gene is 23.9 kb in length. This region is mosaic with areas of high and low GC contents, suggesting that it arose from multiple genetic events. The genetic load region of pNDM-HN380 contained 22 ORFs, of which nine were found in pIncX-SHV. The nine ORFs with high homology in the two plasmids include one resistance gene (blaSHV), three mobile genetic elements (IS26, Tn3 and tnpA) and five ORFs of unknown functions (ΔumuD, ygbI, Δ ygbJ, mpr and orf29). However, pIncX-SHV and pNDM-HN380 had two different alleles of blaSHV that differed from each other by five nucleotides and two amino acids (Gly234 and Glu235 in SHV-11 versus Ser234 and Lys235 in SHV-12).
In pNDM-HN380 (), the blaNDM-1 gene was flanked by ISAba125 and IS26 in the 5′ and 3′ regions, respectively. This 10.8 kb blaNDM-1-containing transposon-like structure was inserted between the truncated ygbj gene (encoding a putative dehydrogenase) and the transponase, Tn3. The ISAba125 element upstream of blaNDM-1 was interrupted by an IS5 element and a 5-bp target site duplication (CCTAA) was identified at the point of insertion between the 5′ end of the IS5 element and the ISAba125 fragment. In the blaNDM-1 upstream region, there was a 3-bp target site duplication (AAC) at the point of insertion between Tn3 and ISAba125 (), suggesting that this was a transposition event. The 3-bp (AAC) target site duplication was identical to that described for pNDM-BJ01 (accession number JQ001791) but different from that for strain 161/07 (accession number HQ857107). No target site duplication repeats could be identified in the sequence adjacent to the IS26 element in the 3′ region. The genes found downstream of blaNDM-1 include the bleomycin resistance gene (bleMBL) and a truncated trpF gene, followed by two ORFs displaying homology (∼70%) with the genome of Stenotrophomonas maltophilia K279a (accession number AM743169), and genes encoding chaperonin subunits (truncated groS and groEL) and the transposase insE. The genetic structure of this transposon (except for the IS26 element in the blaNDM-1 downstream region and the interruption of ISAba125 by IS5), including part of the sequences spanning the junctions (), was identical to those described in the Acinetobacter lowffii plasmid pNDM-BJ01 (INSDC-GenBank accession JQ001791) and in the chromosome of A. baumannii 161/07 (INSDC-GenBank accession HQ857107).
Discussion
The present study revealed the presence of blaNDM-1 in multiple Enterobacteriaceae isolates recovered from returning travelers who have been treated in different parts of China. With the exception of two patients who were of the same family,Citation11 the other patients were not epidemiologically related to each other. Since the isolates were identified by active surveillance upon admission, we concluded that they represent blaNDM-1 importations. In Hong Kong, a territory-wide surveillance for carbapenemases has been implemented since the last quarter of 2008.Citation26 Beside admission screening of at risk patients, microbiology laboratories routinely refer all carbapenem-resistant Enterobacteriaceae (CRE) isolates to a centralized laboratory for molecular testing.Citation26 During the study period, over 500 CRE isolates have been tested by PCR assays. Up to May 2012, a total of 10 blaNDM-1-carrying isolates, including one previously reported by us,Citation26 were identified. The findings suggest that the spread of blaNDM-1 in China is much wider than previously realized. Previous studies have identified blaNDM-1 among clinical isolates of A. baumannii and non-baumannii isolates in Beijing and six provinces (Guangdong, Zhejiang, Hainan, Anhui, Liaoning and Shandong) from patients without history of foreign travelCitation10,Citation12 and in a chicken strain of A. lwoffi in Shandong Province.Citation34 In Hong Kong, the existing strategy only tests patients with a history of recent hospitalization or surgery abroad, those who have traveled to NDM-endemic countries but without hospitalization are not screened.Citation11,Citation26 Given that foreign travel alone has been shown to be an important risk factor for acquisition of antibiotic-resistant enteric bacteria, such as CTX-M producing Enterobacteriaceae,Citation35,Citation36 the number of NDM-positive patients reported here may be an underestimation.
We described here a novel conjugative, blaNDM-1-carrying plasmid in multiple Enterobacteriaceae strains. The findings from the RFLP analysis demonstrated that the IncX3 plasmid has disseminated among multiple enterobacterial species (E. coli, K. pneumoniae, C. freundii and E. cloacae) originating from patients with epidemiological links to multiple geographic areas in China. Since most of the patients had contacts with hospitals, nosocomial dissemination of blaNDM-1 involving the horizontal transfer of the plasmid among hospitals in different areas of China is likely. Recently, two studies have demonstrated limited nosocomial transmission of blaNDM-1 -producing isolates in non-endemic areas.Citation21,Citation37 We previously showed that the two blaNDM-1-carrying E. coli strains (CRE396 and CRE397) in the infant and her mother were clonally related.Citation11 Here, we confirmed that there was in vivo transfer of the blaNDM-1-carrying IncX3 plasmid among E. coli, K. peumoniae and E. aerogenes strains carried by the infant. Although plasmids have been implicated to play a major role in the dissemination of blaNDM-1 in Enterobacteriaceae, the plasmids in the same or different geographic areas either belonged to different incompatibility groups or were different from each other.Citation2,Citation20 Therefore, this is the first time that an epidemic plasmid is implicated in blaNDM-1 dissemination. Since the IncX3 subgroup of family could not be amplified with the initially described PCR-based replicon typing scheme,Citation29 our current understanding of the epidemiology of this group of plasmids is limited. According to the type of associated resistance, genes previously localized on IncX plasmids included: β-lactams (blaTEM-1, blaTEM-52, blaSHV-1) quinolones (qnrS1), amonoglycoisdes (aphA1), olaquindox (oqxAB) and bleomycin (blmS).Citation31 In general, the resistance genes were recruited into a variable genetic load region by IS elements and transposons, while the other plasmid scaffolds were conserved.Citation31 In a collection of 47 E. coli isolates from cases of porcine post-weaning diarrhoea, up to 34% of them were found to be positive for different subgroups of the IncX plasmids not carrying blaNDM-1. Since resistance in food animals could disseminate explosively, future studies should explore possible roles play by animal pathogens and commensal in the dissemination of blaNDM-1.Citation34
This is the first characterized blaNDM-1-carrying IncX3 plasmid, in which the blaNDM-1 was identified inside a composite transposon-like structure flanked by IS26 and ISAba125. It seems that the 10.8 kb blaNDM-1 containing module was integrated en bloc into the IncX3 resistance load region by a recombination event involving IS26 and possibly the other mobile elements flanking the junctions. Our findings were in agreement with the horizontal transfer of the entire module (comprising the ISAba125 fragment with the −35 promoter region, the blaNDM-1 gene, the bleomycin resistance gene, the truncated trpF gene, followed by the tat and dct, the chaperonin subunits, groES and groEL, and the transponase, insE ) from the genus Acinetobacter to Enterobacteriaceae, as suggested previously.Citation10 In Acinetobacter, transposon Tn125 appeared to be the main vehicle for dissemination of blaNDM-1.Citation10,Citation38 This and previous studies indicate that further transfer to Enterobacteriaceae requires other mobile elements, such as IS26 (pMR0211, JN687470; pGUE-NDM, JQ364967; and pNDM-HK, HQ451074), IS903 (pNDM-1_Dok01, AP012208), ISkpn14 (pNDM-KN, JN157804 and pNDM10505, JF503991), IS1 (pNDM10469, JN861072), ISEc33 (p271A, JF785549) and Tn3 (pNDM-MAR, JN420336 and pKpANDM-1, FN396876).Citation8,Citation16,Citation18,Citation19,Citation26,Citation39,Citation40 The IncX plasmids were thought to be narrow host range plasmids of Enterobacteriaceae, but the ability of transfer to Pseudomonas aeruginosa has been demonstrated.Citation41 In the future, it would be interesting to investigate the transferability of IncX plasmids to the genus Acinetobacter which would be expected to facilitate the inter-genera flow of resistance genes.
The backbone of pNDM-HN380 is organized similarly to the backbone of IncX plasmids.Citation31 The tandem genes topB-hns, which act as a conserved stealth module that stabilizes plasmid DNA, is present in all but one (pLN126_33) of the completely sequenced IncX plasmids.Citation31,Citation42 The topB gene is a paralogue of a chromosomally encoded topoisomerase III gene in E. coli.Citation42 In gram-negative bacteria, the H-NS protein is a global repressor of transcription which modulates diverse functions that include biogenesis of flagella and expression of genes acquired horizontally.Citation43 It has been proposed that H-NS binds to curved AT-rich DNA. Therefore, changes in the DNA bend as a result of increase in temperature would weaken the binding, thereby providing a mechanism for dynamic modulation of gene expression in relation to changes in environmental temperature.Citation44 Recently, the plasmid-encoded Sfh protein, which is an H-NS homologue, has been found to allow plasmids to be transmitted to new bacterial hosts with minimal effects on their fitness.Citation45
This study does not have enough data to determine the origin of the blaNDM-1-carrying bacteria with links to China. Those cases had not travelled to the Indian subcontinent, but we cannot exclude the possibility that blaNDM-1-carrying bacteria were acquired from contacts with other people with such travel history. Since the sequences flanking blaNDM-1 in pNDM-HN380 were identical to those having links to the Indian subcontinent, an independent gene escape seems less likely. Nonetheless, it might be speculated that the IncX3 plasmid could be a specific vehicle for blaNDM-1 in China.
In conclusion, this study identified a novel blaNDM-1-carrying IncX3 plasmid disseminated among multiple species of Enterobacteriaceae originating from patients with links to widely separated areas in China. The emergence of NDM-1 in China has likely been contributed by inadequate surveillance, misuse of antimicrobial agents and an incomplete infection control infrastructure in the hospitals. These issues should be addressed as a matter of national healthcare priority. Further studies will be necessary to unveil the full extent of NDM-1 in the country and to investigate the prevalence of this novel plasmid among gram-negative bacteria.
We thank Levina Lam, Ben Ho and Wilson Chan of the Centre for Genomic Sciences, University of Hong Kong for technical assistance. This work was supported by a commission grant (grant NO HK-09-01) from the Research Fund for the Control of Infectious Diseases (RFCID) of the Health and Food Bureau of the Hong Kong Special Administative Region Government. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
- Yong D, Toleman MA, Giske CG et al.Characterization of a new metallo-beta-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother2009;53: 5046–5054.
- Kumarasamy KK, Toleman MA, Walsh TR et al.Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis2010;10: 597–602.
- Walsh TR, Weeks J, Livermore DM, Toleman MA.Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis2011;11: 355–362.
- Bogaerts P, Bouchahrouf W, de Castro RR et al.Emergence of NDM-1-producing Enterobacteriaceae in Belgium. Antimicrob Agents Chemother2011;55: 3036–3038.
- Halaby T, Reuland AE, Al Naiemi N et al.A case of New Delhi metallo-beta-lactamase 1 (NDM-1)-producing Klebsiella pneumoniae with putative secondary transmission from the Balkan region in the Netherlands. Antimicrob Agents Chemother2012;56: 2790–2791.
- Mazzariol A, Bosnjak Z, Ballarini P et al.NDM-1-producing Klebsiella pneumoniae, Croatia. Emerg Infect Dis2012;18: 532–534.
- Poirel L, Schrenzel J, Cherkaoui A, Bernabeu S, Renzi G, Nordmann P.Molecular analysis of NDM-1-producing enterobacterial isolates from Geneva, Switzerland. J Antimicrob Chemother2011;66: 1730–1733.
- McGann P, Hang J, Clifford RJ et al.Complete sequence of a novel 178-kilobase plasmid carrying blaNDM-1 in a Providencia stuartii strain isolated in Afghanistan. Antimicrob Agents Chemother2012;56: 1673–1679.
- Poirel L, Ozdamar M, Ocampo-Sosa AA, Turkoglu S, Ozer UG, Nordmann P.NDM-1-producing Klebsiella pneumoniae now in Turkey. Antimicrob Agents Chemother2012;56: 2784–2785.
- Fu Y, Du X, Ji J, Chen Y, Jiang Y, Yu Y.Epidemiological characteristics and genetic structure of blaNDM-1 in non-baumannii Acinetobacter spp in China. J Antimicrob Chemother2012;67: 2114–2122.
- Ho PL, Li Z, Lai EL, Chiu SS, Cheng VC.Emergence of NDM-1-producing Enterobacteriaceae in China. J Antimicrob Chemother2012;67: 1553–1555.
- Hu H, Hu Y, Pan Y et al.Novel plasmid and its variant harboring both a blaNDM-1 gene and type IV secretion system in clinical isolates of Acinetobacter lwoffii. Antimicrob Agents Chemother2012;56: 1698–1702.
- Girlich D, Poirel L, Nordmann P.Diversity of naturally occurring Ambler class B metallo-beta-lactamases in Erythrobacter spp. J Antimicrob Chemother;e-pub ahead of print 31 July 2012; https://doi.org/10.1093/jac/dks289 doi:10.1093/jac/dks289
- Zheng B, Tan S, Gao J et al.An unexpected similarity between antibiotic-resistant NDM-1 and beta-lactamase II from Erythrobacter litoralis. Protein Cell2011;2: 250–258.
- Derzelle S, Duchaud E, Kunst F, Danchin A, Bertin P.Identification, characterization, and regulation of a cluster of genes involved in carbapenem biosynthesis in Photorhabdus luminescens. Appl Environ Microbiol2002;68: 3780–3789.
- Sekizuka T, Matsui M, Yamane K et al.Complete sequencing of the blaNDM-1-positive IncA/C plasmid from Escherichia coli ST38 isolate suggests a possible origin from plant pathogens. PLoS ONE2011;6: e25334.
- Walsh TR.Emerging carbapenemases: a global perspective. Int J Antimicrob Agents2010;36(Suppl 3): S8–S14.
- Villa L, Poirel L, Nordmann P, Carta C, Carattoli A.Complete sequencing of an IncH plasmid carrying the blaNDM-1, blaCTX-M-15 and qnrB1 genes. J Antimicrob Chemother2012;67: 1645–1650.
- Poirel L, Bonnin RA, Nordmann P.Analysis of the resistome of a multidrug-resistant NDM-1-producing Escherichia coli strain by high-throughput genome sequencing. Antimicrob Agents Chemother2011;55: 4224–4229.
- Nordmann P, Poirel L, Walsh TR, Livermore DM.The emerging NDM carbapenemases. Trends Microbiol2011;19: 588–595.
- Kim MN, Yong D, An D et al.Nosocomial clustering of NDM-1-producing Klebsiella pneumoniae sequence type 340 strains in four patients at a South Korean tertiary care hospital. J Clin Microbiol2012;50: 1433–1436.
- Cohen SJ, Leverstein-Van Hall MA.Guideline for phenotypic screening and confirmation of carbapenemases in Enterobacteriaceae. Int J Antimicrob Agents2010;36: 205–210.
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing: twenty-first informational supplement M100-S21. Wayne, PA:CLSI, 2011.Available at http://www.rsu.ac.th/medtech/files/CLSI%202011.pdf
- Endimiani A, Carias LL, Hujer AM et al.Presence of plasmid-mediated quinolone resistance in Klebsiella pneumoniae isolates possessing blaKPC in the United States. Antimicrob Agents Chemother2008;52: 2680–2682.
- Poirel L, Heritier C, Tolun V, Nordmann P.Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother2004;48: 15–22.
- Ho PL, Lo WU, Yeung MK et al.Complete sequencing of pNDM-HK encoding NDM-1 carbapenemase from a multidrug-resistant Escherichia coli strain isolated in Hong Kong. PLoS One2011;6: e17989.
- Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S.Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol2005;43: 4178–4182.
- Wirth T, Falush D, Lan R et al.Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol2006;60: 1136–1151.
- Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ.Identification of plasmids by PCR-based replicon typing. J Microbiol Methods2005;63: 219–228.
- Villa L, Garcia-Fernandez A, Fortini D, Carattoli A.Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother2010;65: 2518–2529.
- Johnson TJ, Bielak EM, Fortini D et al.Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid2012;68: 43–50.
- Aziz RK, Bartels D, Best AA et al.The RAST Server: rapid annotations using subsystems technology. BMC Genomics2008;9: 75.
- Rissman AI, Mau B, Biehl BS, Darling AE, Glasner JD, Perna NT.Reordering contigs of draft genomes using the Mauve aligner. Bioinformatics2009;25: 2071–2073.
- Wang Y, Wu C, Zhang Q et al.Identification of New Delhi metallo-beta-lactamase 1 in Acinetobacter lwoffii of food animal origin. PLoS ONE2012;7: e37152.
- Tangden T, Cars O, Melhus A, Lowdin E.Foreign travel is a major risk factor for colonization with Escherichia coli producing CTX-M-type extended-spectrum beta-lactamases: a prospective study with Swedish volunteers. Antimicrob Agents Chemother2010;54: 3564–3568.
- Kennedy K, Collignon P.Colonisation with Escherichia coli resistant to “critically important” antibiotics: a high risk for international travellers. Eur J Clin Microbiol Infect Dis2010;29: 1501–1506.
- Denis C, Poirel L, Carricajo A et al.Nosocomial transmission of NDM-1-producing Escherichia coli within a non-endemic area in France. Clin Microbiol Infect2012;18: E128–E130.
- Poirel L, Bonnin RA, Boulanger A, Schrenzel J, Kaase M, Nordmann P.Tn125-related acquisition of blaNDM-like genes in Acinetobacter baumannii. Antimicrob Agents Chemother2012;56: 1087–1089.
- Bonnin RA, Poirel L, Carattoli A, Nordmann P.Characterization of an IncFII plasmid encoding NDM-1 from Escherichia coli ST131. PLoS ONE2012;7: e34752.
- Carattoli A, Villa L, Poirel L, Bonnin RA, Nordmann P.Evolution of IncA/C blaCMY-2-carrying plasmids by acquisition of the blaNDM-1 carbapenemase gene. Antimicrob Agents Chemother2012;56: 783–786.
- Tardif G, Grant RB.Transfer of plasmids from Escherichia coli to Pseudomonas aeruginosa: characterization of a Pseudomonas aeruginosa mutant with enhanced recipient ability for enterobacterial plasmids. Antimicrob Agents Chemother1983;24: 201–208.
- Norman A, Hansen LH, She Q, Sorensen SJ.Nucleotide sequence of pOLA52: a conjugative IncX1 plasmid from Escherichia coli which enables biofilm formation and multidrug efflux. Plasmid2008;60: 59–74.
- Ko M, Park C.Two novel flagellar components and H-NS are involved in the motor function of Escherichia coli. J Mol Biol2000;303: 371–382.
- Dorman CJ.H-NS, the genome sentinel. Nat Rev Microbiol2007;5: 157–161.
- Doyle M, Fookes M, Ivens A, Mangan MW, Wain J, Dorman CJ.An H-NS-like stealth protein aids horizontal DNA transmission in bacteria. Science2007;315: 251–252.