Abstract
Pyrazinamide (PZA) is a frontline anti-tuberculosis drug that plays a crucial role in the treatment of both drug susceptible and multidrug-resistant tuberculosis (MDR-TB). Resistance to PZA is most commonly associated with mutations in the pncA gene encoding nicotinamidase/pyrazinamidase which converts the prodrug PZA to the active form pyrazinoic acid (POA). RpsA (ribosomal protein S1) involved in trans-translation was recently shown to be a target of PZA and mutations in RpsA are found in some PZA-resistant TB strains. However, some other PZA-resistant strains lack mutations in either pncA or rpsA. To identify potential new mechanisms of PZA resistance, we isolated 174 in vitro mutants of M. tuberculosis H37Rv resistant to PZA to search for resistant isolates that do not have pncA or rpsA mutations. DNA sequencing revealed that 169 of the 174 (97.1%) PZA-resistant mutants had pncA mutations but 5 mutants lacked pncA or rpsA mutations. Whole genome sequencing analyses revealed that the 5 PZA-resistant mutants had different mutations all occurring in the same gene panD encoding aspartate decarboxylase, which is involved in synthesis of β-alanine that is a precursor for pantothenate and co-enzyme A biosynthesis. panD mutations were identified in naturally PZA-resistant Mycobacterium canetti strain and a PZA-resistant MDR-TB clinical isolate. Future studies are needed to address the role of panD mutations in PZA resistance and confirm PanD as a new target of PZA.
Emerging Microbes & Infections (2013) 2, e35; doi:10.1038/emi.2013.38
Introduction
Pyrazinamide (PZA) is an important first-line tuberculosis (TB) drug used in combination with other TB drugs for the treatment of both drug susceptible TB and multidrug-resistant tuberculosis MDR-TB.Citation1 PZA plays a critical role in modern TB chemotherapyCitation2 by shortening the treatment from previously 9-12 months to 6 months. The unique sterilizing activity of PZA in shortening the treatment is due to its high activity against persister bacteria that are not killed by other TB drugs.Citation3 Because of its indispensible sterilizing activity, all new TB drug candidates in clinical trials are used together with PZA.Citation4,Citation5,Citation6,Citation7
PZA is an unconventional and paradoxical TB drug, characterized by lack of activity against growing bacteria under normal culture conditions,Citation8 but high activity for non-replicating persisters at acid pH (e.g. pH 5.5).Citation9 Even under acid pH conditions in vitro, PZA has a high minimal inhibitory concentration (MIC) of 50–100 µg/mL at pH 5.5–6.0. PZA is a peculiar drug whose activity is influenced by various factors such as acid pH, culture age, starvation, weak acids, energy inhibitors and microaeropHilic/anaerobic conditions. Despite the importance of PZA in shortening the treatment of TB, its mechanism of action is the least understood of all TB drugs. Structurally, PZA is an analog of nicotinamide. Mutation in pncA encoding nicotinamidase/pyrazinamidase (PZase)Citation10 is the major mechanism for PZA resistance in M. tuberculosis.Citation10,Citation11,Citation12 Like isoniazid,Citation13 PZA is a prodrug which requires activation to its active form pyrazinoic acid (POA) by M. tuberculosis PZase enzyme.Citation10 Recently, we identified a new target of PZA as ribosomal protein S1 (RpsA, Rv1630), a vital ribosomal protein involved in trans-translation.Citation14 Trans-translation is involved in degradation of potentially toxic protein products formed in stressed bacteria required for persister survival. Mutations in rpsA have been found in some PZA-resistant strains without pncA mutations.Citation14,Citation15,Citation16 However, some PZA-resistant strains, which are typically low level PZA resistant (MIC=200–300 µg/mL, PH 6.0) and PZase positive do not have mutations in either pncA or the rpsA gene.Citation12,Citation15,Citation17 To identify new mechanisms of PZA resistance, in this study, we isolated a large number of in vitro generated mutants resistant to PZA and characterized these strains for novel mutations in their genomes by whole genome sequencing. Sequence analyses of 5 low level PZA-resistant isolates without pncA or rpsA mutations indicate mutations in the panD gene encoding aspartate alpha-decarboxylase as a potential new mechanism of PZA resistance.
MATERIALS AND METHODS
Isolation of M. tuberculosis mutants resistant to PZA and PZA susceptibility testing
Mycobacterium tuberculosis H37Rv was grown in 7H9 liquid medium (Difco) supplemented with 0.05% Tween 80 and 10% bovine serum albumin-dextrose-catalase (ADC) enrichment at 37 °C for approximately 10–14 days (mid- to late-exponential phase) with occasional agitation as described.Citation10 Pyrazinamide (Sigma-Aldrich Co.) was dissolved in deionized water at a stock concentration of 10 mg/mL and filter-sterilized and incorporated into 7H11 agar plates containing ADC at concentrations of 200 µg/mL, pH 6.0. Mutants that grew on the PZA containing plates after 3–4 weeks incubation at 37 °C were picked and grown in 7H9 liquid medium for confirming PZA resistance pHenotype by repeated PZA susceptibility testing. The PZA susceptibility testing of the PZA-resistant mutants was performed on 7H11 agar plates containing 100 µg/mL, 200 µg/mL, 300 µg/mL PZA (pH6.0) as described.Citation18 Wild type M. tuberculosis H37Rv and a known PZA-resistant mutant PZA-R1 containing a pncA mutation (Q10P) were included as a drug susceptible control strain and a resistant control strain for the PZA susceptibility testing. PZA susceptible strain H37Rv did not grow on PZA containing plates while the PZA-resistant mutants and PZA-R1 resistant control strain grew on PZA containing plates.
Polymerase chain reaction (PCR) and DNA sequencing
The pncA PCR was performed using P1 primer (5-GTCGGTCATGTTCGCGATCG-3; from -105 base pair (bp) upstream of pncA) and P6 primer (5-GCTTTGCGGCGAGCGCTCCA-3; from 60 base pair downstream of the stop codon) as described.Citation11 Briefly, genomic DNA from 175 in vitro isolated PZA-resistant mutants was isolated (see below) and used as templates for PCR as follows: heat denaturation at 94 °C 15 min followed by 30 cycles of 94 °C 0.5 min, 55 °C 0.5 min, 72 °C 1 min followed by extension at 72 °C for 7 min. The PCR reaction was then cooled to 4 °C. The pncA PCR products were then sequenced by ABI 377 DNA sequencer at Johns Hopkins Genetic Resources Core Facility, and the pncA sequences from different mutant isolates were compared against the wild type pncA sequence of M. tuberculosis H37Rv to identify potential mutations in the pncA gene. The rpsA gene was also PCR amplified, and the PCR products were sequenced for 5 mutants without pncA mutations using primers and conditions as previously described.Citation14 Primers (panD_F: 5'TCAACGGTTCCGGTCGGCTGCT3' and panD_R: 5'TATCCGCCACTGCTGCACGACCTT3') were used to amplify a 650 bp PCR product that contains the whole panD gene from PZA-resistant M. tuberculosis strains using the same condition as above for amplifying the pncA gene. The 650 bp panD PCR products were sequenced as above to identify possible mutations in panD.
PZase activity determination
The PZase enzyme test (the Wayne PZase test) was performed as described,Citation19 with the following modifications. Briefly, PZA was added to 100 µg/mL final concentration to 1 mL M. tuberculosis log phase cultures in Eppendorf tubes and incubated at 37 °C overnight, and then 2% ferrous sulfate was added for color development. PZA in the presence of positive PZase enzyme from the M. tuberculosis will be converted to POA, which then reacts with ferrous ion to produce a brown colored compound, which can be detected as an indication of positive PZase activity.
Whole genome sequencing
The genomic DNA for whole genome sequencing was isolated as previously described.Citation20 The genomic DNA samples from the 5 PZA-resistant mutants that were positive for PZase and did not have pncA mutations were subjected to whole genome sequencing using Illumina HiSeq 2000 machine. Paired-end sequencing libraries for genomic DNA of each strain were barcoded and constructed with insert sizes of approximately 300 bp using TruSeq DNA Sample Preparation kits (Illumina, USA) according to manufacturer's instruction. For each strain, 1.0 G-1.5 G bases (230-fold to 350-fold genome coverage) were generated after barcodes were trimmed. High-quality data were aligned with the reference sequence of M. tuberculosis H37Ra (NC_009525) using SOAPaligner.Citation21 We used M. tuberculosis H37Ra genome sequenceCitation20 as a reference strain for sequence comparison with the PZA-resistant mutants derived from M. tuberculosis H37Rv because of the significant number of sequencing errors in the original H37Rv genome sequence in the database.Citation22 Only reads where both ends aligned to the reference sequence were used for single nucleotide variant (SNV) and insertion and deletion (InDels) analysis. SNVs and InDels ranging from 1 to 5 bp were sorted and called at minimum reads of 10. In order to eliminate the genomic differences of H37Ra and H37Rv in our analysis, SNVs and InDels shared between H37Ra and H37Rv were further filtered and annotated for gene locus and mutation types with the nearest coding sequences. Synonymous mutations and PE/PPE mutations within coding sequence were not included in the final analysis to focus on mutations that are most likely involved in PZA resistance.
RESULTS
Isolation of PZA-resistant mutants
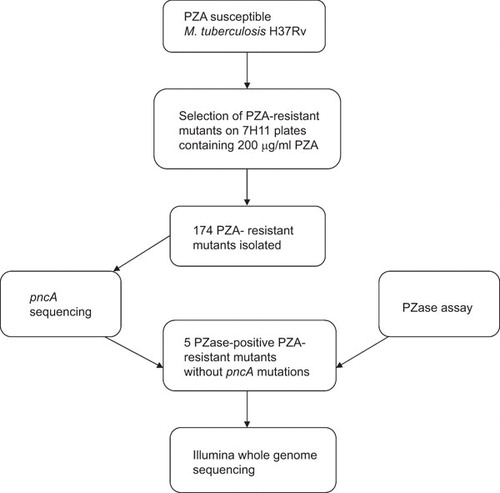
The flow chart of isolation and characterization of in vitro mutants resistant to PZA is shown in . The wild type M. tuberculosis strain H37Rv was susceptible to 100 µg/mL PZA (pH 5.9). To isolate PZA-resistant spontaneous mutants, early stationary phase cultures of M. tuberculosis H37Rv were plated on 7H11 agar plates containing 200 µg/mL PZA (pH 5.9). Through several rounds of isolation, about 300 mutants were obtained. After repeated PZA susceptibility testing to rule out false resistance, a total of 174 mutants were obtained that were consistently resistant to PZA. These 174 PZA-resistant mutants were subjected to further analysis by pncA sequencing as below.
pncA sequencing revealed new PZA-resistant mutants without pncA mutations
To identify desired mutants that do not have pncA mutations which would indicate possible new mechanisms of PZA resistance, we isolated genomic DNA from the 174 PZA-resistant mutants and performed PCR to amplify the pncA gene. DNA sequencing analysis of the pncA PCR products revealed that 169 of the 174 (97.1%) PZA-resistant mutants had various pncA mutations while 5 mutants, S6, S9, S10, S11, S13, did not have any pncA mutations. Sequencing analysis of rpsA, another gene involved in PZA resistance,Citation14 did not show any rpsA mutations in the 5 mutants without pncA mutations. PZase assay showed that the 5 mutants were positive for the enzyme activity, which is consistent with the above pncA sequencing results and also ruled out a pncA promoter or regulatory mutation that could result in lack of PZase enzyme activity as a possible cause of the PZA resistance in the 5 mutants. The above findings suggest that the 5 PZA-resistant mutants harbor possible new mechanisms of PZA resistance independent of pncA or rpsA mutations.
Whole genome sequencing identified a new gene panD closely associated with PZA resistance
To identify possible new mechanisms of PZA resistance, we subjected the 5 PZA-resistant mutants without pncA or rpsA mutations to whole genome sequencing using Illumina Hi-Seq2000. After filtering out PE/PPE family genes and the genomic differences between H37Ra and H37Rv, only 3, 2, 4, 5 and 3 SNVs were identified respectively for mutants S6, S9, S10, S11 and S13, and only 1 InDel was identified for mutants S6, S9 and S10, respectively (Supplementary information Table 1). Comparative genome sequence analyses of the 5 PZA-resistant strains revealed that they all had mutations in a single gene, panD, encoding aspartate alpha-decarboxylase (). It is interesting to note that the 5 mutants had 5 different mutations in the panD gene. Mutant S6 had an A128S mutation (Ala to Ser change at amino acid position 128), S9 and S10 had identical panD mutation V138A, S11 had two mutations causing H21R and I49V substitutions, S13 had an E130G substitution in the panD gene. These 5 mutations revealed by whole genome sequencing were confirmed to be genuine by PCR sequencing of the panD gene from each of the 5 individual mutants.
Table 1 panD mutations identified in PZA-resistant mutants or clinical isolates
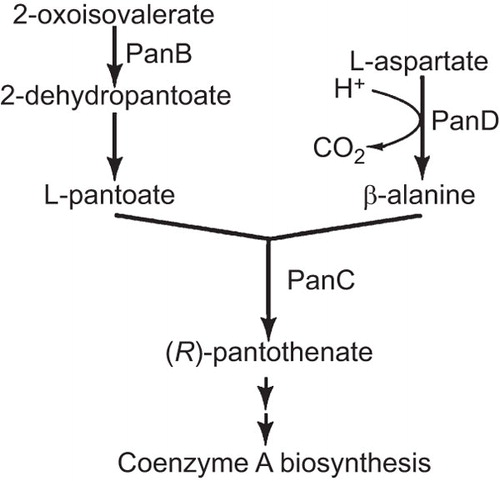
panD is located in an operon lysS-Rv3603c with panC (pantothenate synthetase), Rv3603c (conserved hypothetical alanine and leucine rich protein), Rv3600c (hypothetical protein), Rv3559c (hypothetical protein), and lysS (lysyl-tRNA synthetase 1) (). panD encodes a 139 amino acid (15 kD) protein involved in synthesis of β−alanine from decarboxylation of L-aspartate required for pantothenate (vitamin B5) and co-enzyme A (CoA) biosynthesis ().
Identification of panD mutations in clinical isolates
Mycobacterium canetti, a member of the M. tuberculosis complex that causes human TB in some regions of Africa, is naturally resistant to PZA but lacks pncA mutations.Citation23 It is interesting to note that it contained a non-synonymous mutation of T to C change at nucleotide position 350 causing M117T change and a silent mutation (C39G) in panD (). In addition, an MDR-TB clinical isolate resistant to PZA was found to harbor a mutation of T to C change at nucleotide position 400 causing amino acid substitution of P134S ().
DISCUSSION
In this study, we found that mutations in panD are closely associated with PZA resistance.
PZA resistance in M. tuberculosis is most commonly caused by mutations in pncA geneCitation10 encoding the PZase required for conversion of PZA prodrug to POA or occasionally caused by mutations in the target rpsA encoding ribosomal protein S1.Citation14 However, there are some PZA-resistant clinical isolates that do not have mutations in pncA or rpsA. It has been challenging to pin down the new mechanism of PZA resistance using clinical isolates because of the diverse genetic background of the clinical strains that differ from each other and from the sequenced type strains. By using whole genome sequencing of isogenic mutants from the same strain H37Rv, we were able to identify mutation of the panD gene as a possible new mechanism of PZA resistance. Besides the in vitro isolated mutants that have panD mutations, we also found panD mutations in clinical isolates such as M. canettii and a clinical strain. Although M. canettii which is naturally resistant to PZA was recently found to harbor rpsA mutations,Citation16 it is worth noting that M. canettii also had an M117T amino acid substitution in the PanD. The relative contribution of the rpsA and panD mutations in the natural PZA resistance of M. canettii remains to be determined. The finding that panD mutations are closely associated with PZA resistance may offer yet a third mechanism of PZA resistance besides pncA and rpsA mutations. However, there may be other unidentified genes involved in PZA resistance, since we found that the PZA-resistant clinical isolate 9739 (PZA MIC=200–300 µg/mL)Citation12 does not have any mutations in pncA, rpsA, or panD (data not shown).
panD mutation in M. tuberculosis has been shown to cause higher attenuation of virulence in mice than BCG vaccine,Citation24 indicating panD may be critical for survival and persistence of the bacilli in vivo. panD encoding aspartate alpha-decarboxylase is involved in synthesis of β−alanine which is in turn required for pantothenate and CoA synthesis. CoA has a central role in cellular metabolism. CoA is similar to nicotinamide adenine dinucleotide and flavin adenine dinucleotide (FAD) in structure and serves as an acetyl group carrier important for synthesis and oxidation of fatty acids and oxidation of pyruvate in the Tricarboxylic acid cycle to generate ATP. The possibility that PZA may inhibit pantothenate and CoA synthesis thereby interfering with diverse metabolic functions such as energy production and fatty acid metabolism in M. tuberculosis needs to be addressed in future studies.
Although a few other mutations such as mutations in HadC (β-hydroxyacyl- acyl carrier protein dehydratase) involved in cell wall mycolic acid elongation were identified in 3 of the 5 PZA-resistant mutants (Supplementary information Table 1), they are less likely causal in PZA-resistance. This is because mycolic acid synthesis mainly occurs in growing TB bacteria and inhibition by PZA of HadC responsible for mycolic acid elongation, while cannot be excluded, is inconsistent with the unique activity of PZA for non-growing persisters. Nevertheless, future studies are required to rule out the possibility of HadC mutations being involved in PZA resistance.
In summary, we identified a new gene panD whose mutations are closely associated with PZA resistance in PZA-resistant mutants and clinical isolates without pncA or rpsA mutations. panD may encode another target of PZA in addition to RpsA. Future studies are needed to assess the role of the identified panD mutations as a new mechanism of PZA resistance and confirm the role of PanD as a new target of PZA in M. tuberculosis.
Figure 3 PanD is involved in synthesis of β-alanine, which is a precursor for pantothenate and coenzyme A biosynthesis in M. tuberculosis. Enzymes involved in the biosynthesis of pantothenate in M. tuberculosis include: PanB, ketopantoate hydroxymethyl transferase; PanD, L-aspartate alpha-decarboxylase; PanC, pantothenate synthetase.
Supplementary Figure 1
Download MS Excel (11.9 KB)The work was supported in part by NIH grant AI099512 and Major Project of the Twelfth Five-Year Plan (2013ZX10003008-003 and 2013ZX10003001-002).
- World Health Organization. Treatment of Tuberculosis: Guidelines, Fourth edition.Geneva: World Health Organization Press, 2010.
- Zhang Y, Mitchison D.The curious characteristics of pyrazinamide: a review. Int J Tuberc Lung Dis2003;7: 6–21.
- Mitchison DA.The action of antituberculosis drugs in short course chemotherapy. Tubercle1985;66: 219–225.
- Andries K, Verhasselt P, Guillemont J et al.A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science2005;307: 223–227.
- Rosenthal IM, Zhang M, Williams KN et al.Daily dosing of rifapentine cures tuberculosis in three months or less in the murine model. PLoS Med2007;4: e344.
- Tasneen R, Li SY, Peloquin CA et al.Sterilizing Activity of Novel TMC207- and PA-824-Containing Regimens in a Murine Model of Tuberculosis. Antimicrob Agents and Chemothe2011;55: 5485–5492.
- Tasneen R, Tyagi S, Williams K, Grosset J, Nuermberger E.Enhanced bactericidal activity of rifampin and/or pyrazinamide when combined with PA-824 in a murine model of tuberculosis. Antimicrob Agents Chemother2008;52: 3664–3668.
- Tarshis MS, Weed WA Jr.Lack of significant in vitro sensitivity of Mycobacterium tuberculosis to pyrazinamide on three different solid media. Am Rev Tuberc1953;67: 391–395.
- McDermott W, Tompsett R.Activation of pyrazinamide and nicotinamide in acidic environment in vitro. Am Rev Tuberc1954;70: 748–754.
- Scorpio A, Zhang Y.Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat Med1996;2: 662–667.
- Scorpio A, Lindholm-Levy P, Heifets L et al.Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother1997;41: 540–543.
- Cheng SJ, Thibert L, Sanchez T, Heifets L, Zhang Y.pncA mutations as a major mechanism of pyrazinamide resistance in Mycobacterium tuberculosis: spread of a monoresistant strain in Quebec, Canada. Antimicrob Agents Chemother2000;44: 528–532.
- Zhang Y, Heym B, Allen B, Young D, Cole S.The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature1992;358: 591–593.
- Shi W, Zhang X, Jiang X et al.Pyrazinamide inhibits trans-translation in Mycobacterium tuberculosis. Science2011;333: 1630–1632.
- Simons SO, Mulder A, van Ingen J, Boeree MJ, van Soolingen D.Role of rpsA Gene Sequencing in Diagnosis of Pyrazinamide Resistance. J Clin Microbiol2013;51: 382.
- Feuerriegel S, Koser CU, Richter E, Niemann S.Mycobacterium canettii is intrinsically resistant to both pyrazinamide and pyrazinoic acid. J Antimicrob Chemother2013;68:1439–1440.
- Alexander DC, Ma JH, Guthrie JL, Blair J, Chedore P, Jamieson FB.Gene sequencing for routine verification of pyrazinamide resistance in Mycobacterium tuberculosis: a role for pncA but not rpsA. J Clin Microbiol2012;50: 3726–3728.
- Zhang Y, Permar S, Sun Z.Conditions that may affect the results of susceptibility testing of Mycobacterium tuberculosis to pyrazinamide. J Med Microbiol2002;51: 42–49.
- Wayne LG.Simple pyrazinamidase and urease tests for routine identification of mycobacteria. Am Rev Respir Dis1974 ; 109: 147–151.
- Zheng H, Lu L, Wang B et al.Genetic basis of virulence attenuation revealed by comparative genomic analysis of Mycobacterium tuberculosis strain H37Ra versus H37Rv. PLoS One2008;3: e2375.
- Li R, Li Y, Kristiansen K, Wang J.SOAP: short oligonucleotide alignment program. Bioinformatics2008;24: 713–714.
- Ioerger TR, Feng Y, Ganesula K et al.Variation among genome sequences of H37Rv strains of Mycobacterium tuberculosis from multiple laboratories. J Bacteriol2010;192: 3645–3653.
- Somoskovi A, Dormandy J, Parsons LM et al.Sequencing of the pncA gene in members of the Mycobacterium tuberculosis complex has important diagnostic applications: Identification of a species-specific pncA mutation in "Mycobacterium canettii" and the reliable and rapid predictor of pyrazinamide resistance. J Clin Microbiol2007;45: 595–599.
- Sambandamurthy VK, Wang X, Chen B et al.A pantothenate auxotroph of Mycobacterium tuberculosis is highly attenuated and protects mice against tuberculosis. Nat Med2002;8: 1171–1174.