Abstract
To evaluate the association between mutations in the genes gyrA/B and resistance levels to fluoroquinolones in clinical isolates of Mycobacterium tuberculosis, a total of 80 ofloxacin-resistant isolates collected in 2009 by the Shanghai Municipal Centers for Disease Control and Prevention were studied. The minimum inhibitory concentration (MIC) of ofloxacin, moxifloxacin and gatifloxacin for each isolate was determined using the microscopic observation drug susceptibility assay. Sequencing was used to identify mutations in the quinolone resistance-determining region (QRDR) of the gyrA and gyrB genes. In total, 68 isolates had mutations in gyrA, three isolates had mutations in gyrB, six isolates had mutations in both gyrA and gyrB, and three isolates had no mutations. Two common mutations in gyrA, the D94G and D94N mutations, were associated with higher-level resistance to all three fluoroquinolones than two other common mutations (A90V and D94A). Understanding the relationship between MICs and mutations in ofloxacin-resistant isolates will facilitate the optimization of the use of new-generation fluoroquinolones to treat patients with ofloxacin-resistant tuberculosis (TB).
Introduction
Fluoroquinolones, including the older-generation drug, ofloxacin, and the newer-generation drugs, gatifloxacin and moxifloxacin, are second-line anti-tuberculosis agents.Citation1 These agents are used in the treatment of drug-resistant tuberculosis, particularly for multidrug-resistant tuberculosis (TB), which is defined as resistance to at least isoniazid and rifampicin.Citation1 Fluoroquinolones inactivate Mycobacterium tuberculosis by binding to gyrase-DNA complexes and inhibiting DNA replication.Citation2 The widespread and erroneous use of fluoroquinolones in TB treatment has led to the recent emergence of resistance.Citation3
M. tuberculosis acquires resistance to fluoroquinolones mainly through mutations in conserved regions, referred to as quinolone resistance-determining regions (QRDRs), of the gyrA and gyrB genes, which encode DNA gyrase.Citation2,Citation4,Citation5 DNA gyrase contains a drug-binding pocket called the quinolone-binding pocket (QBP), which consists of both amino acid residues and DNA nucleotides.Citation6,Citation7 Mutations in the two genes change the structure of the QBP and may lead to broad cross-resistance to all fluoroquinolones.Citation8,Citation9 The most frequent mutations in clinical isolates are found at codons 90 (A90V), 91 (S91P) and 94 (D94G, D94A, D94N and D94Y) of gyrA.Citation10,Citation11,Citation12 Mutations of codons 500, 538, 539 and 540 in gyrB are also related to resistance to fluoroquinolones.Citation13,Citation14 The altered amino acids encoded by these gene mutations are all located within the QBP and interact directly with quinolones.Citation9,Citation13 Because every generation of fluoroquinolones has the same drug targets, cross-resistance among fluoroquinolones is common. However, the resistance levels of each isolates against individual drugs are variable. In general, the minimum inhibitory concentrations (MICs) of newer generation fluoroquinolones are much lower than those of older generation fluoroquinolones.Citation15 Thus, the newer-generation fluoroquinolones are reported to retain efficacy in the treatment of ofloxacin-resistant cases.Citation15,Citation16,Citation17
Despite current knowledge, the association between resistance levels and different mutations remains debatable. Some studies have shown that mutations in codon 94 in gyrA are related to higher-level resistance, whereas mutations in codon 91 of gyrA may be associated with lower-level resistance.Citation10,Citation11,Citation12 However, a study by Cui and colleagues suggested that there may be no significant association between gyrA mutations and the levels of resistance to ofloxacin.Citation18 Thus, this association is unclear and requires further exploration. As molecular techniques have been increasingly applied to the rapid diagnosis of drug resistant M. tuberculosis,Citation19 clarifying the fluoroquinolone resistance levels of M. tuberculosis that carry different gyrA and gyrB mutations will help optimize therapeutic regimens in clinical TB treatment.
To explore the association between resistance levels to fluoroquinolones and different mutations in M. tuberculosis, we determined the MICs of ofloxacin, moxifloxacin and gatifloxacin on 80 ofloxacin-resistant isolates and identified the mutations in gyrA/B.
MATERIALS AND METHODS
Bacterial isolates
A total of 1960 M. tuberculosis isolates were collected by the Shanghai Municipal Centers for Disease Control and Prevention in 2009. The identification of these M. tuberculosis isolates was performed by conventional biochemical and polymerase chain reaction (PCR) tests.Citation20 Among the isolates, 458 were identified as phenotypically resistant to at least one first-line anti-TB drug (isoniazid, rifampicin, ethambutol or streptomycin) by the proportion method on Lowenstein–Jensen medium with the following concentrations: isoniazid (0.2 mg/L), rifampin (40 mg/L), ethambutol (2 mg/L) and streptomycin (4 mg/L). The susceptibility of these 458 isolates to ofloxacin was further tested at a breakpoint concentration of 2 mg/L on Lowenstein–Jensen medium using the proportion method. From these analyses, 80 ofloxacin-resistant isolates were identified and selected for further study. M. tuberculosis H37Rv (ATCC 27294) was included as the reference strain.
Determination of MICs of ofloxacin, moxifloxacin and gatifloxacin
The MICs of ofloxacin (Sigma-Aldrich Co., St Louis, MO, USA), moxifloxacin (TRC Co., Toronto, Canada) and gatifloxacin (Sigma-Aldrich Co.) for the 80 isolates were determined using the microscopic observation drug susceptibility assay as recommended at http://www.modsperu.org/.Citation21,Citation22 The growth of M. tuberculosis was observed microscopically as the formation of characteristic strings and tangles. The validity of microscopic observation drug susceptibility for determining fluoroquinolone resistance has been demonstrated previously.Citation22 Ofloxacin was tested at concentrations of 0.5, 1.0, 2.0, 4.0, 8.0, 16.0 and 32.0 mg/L; gatifloxacin and moxifloxacin were tested at concentrations of 0.125, 0.25, 0.5, 1.0, 2.0, 4.0 and 8.0 mg/L. MICs were determined visually as the minimum drug concentration that prevented the growth of cord structures. The cutoff concentrations of resistance to ofloxacin, moxifloxacin and gatifloxacin were 2.0 mg/L, 0.5 mg/L and 0.5 mg/L, respectively.Citation22,Citation23
DNA extraction
A loopful of M. tuberculosis culture from Lowenstein–Jensen medium was transferred to a 1.5 mL tube that contained 400 µL of TB lysis buffer (10 mmol/L Tris-HCl, 1 mmol/L EDTA (pH 8.0)) and heat inactivated at 85 °C for 30 min. The tube was then centrifuged at 10 000 g at 4 °C for 10 min. The supernatant was transferred to a fresh 1.5-mL tube and stored at −20 °C until use.
Sequencing of the QRDRs of the gyrA and gyrB genes
The primers for PCR amplification were newly designed using gyrA and gyrB sequences (GenBank accession number L27512) encompassing the QRDRs of these two genes (forward, 5′-CAG CTA CAT CGA CTA TGC GA-3′ and reverse, 5′-GGG CTT CGG TGT ACC TCA T-3′ for gyrA; forward, 5′-CGT AAG GCA CGA GAG TTG GT-3′ and reverse, 5′-ATC TTG TGG TAG CGC AGC TT-3′ for gyrB). The basic 50-µL amplification reaction mixture contained 3 µL DNA, 20 µL 2×PCR MIX (TianGen, Beijing, China), 10 µM PCR primer mixture (Sangon, Shanghai, China) and 17 µL distilled water. The following steps were performed: one denaturation cycle at 94 °C for 10 min, followed by 30 cycles of 45 s at 94 °C, 30 s at 56 °C and 50 s at 72 °C, followed by elongation at 72 °C for 10 min. Both PCR products were 320 base pairs in size. They were purified and sequenced by Sunnybio (Shanghai, China). Mutations of gyrA and gyrB were identified by aligning sequences of resistant isolates to the sequence of M. tuberculosis strain H37Rv using CLUSTAL W (European Bioinformatics Institute).
Statistical analysis
The association of gyrA mutations with MIC levels was analyzed using the Kruskal–Wallis test and Mann–Whitney U test in the Statistical Package for Social Sciences version 12.0 software (SPSS Inc., Chicago, IL, USA).
Results
To study the association between mutations and resistance levels, we determined the MICs of the three fluoroquinolones for the M. tuberculosis isolates and identified associated mutations in the gyrA/B genes. The profiles of these isolates () show that the MIC50 values of ofloxacin, moxifloxacin and gatifloxacin was 4 mg/L, 1 mg/L and 0.5 mg/mL, respectively. Seventy-seven isolates (96.25%, 77/80) carried mutations in the QRDRs of gyrA/B (); of those, 68 (88.3%, 68/77) had mutations in gyrA (at codons 88, 89, 90, 91 and 94); three isolates had mutations in gyrB; and the remaining six isolates had mutations in both the gyrA and gyrB genes. Based on the cutoff concentrations used, only one isolate carried no mutation in gyrA or gyrB and showed susceptibility to gatifloxacin and resistance to ofloxacin and moxifloxacin. All of the other isolates had broad cross-resistance to all three of the drugs.
Table 1 The mutation profiles and the fluoroquinolone MICs of the 80 ofloxacin-resistant M. tuberculosis isolates
A total of 61 (76.25%, 61/80) isolates carried the four most frequent mutations in gyrA: A90V (n=18), D94A (n=13), D94G (n=18) and D94N (n=12). The MIC distributions of fluoroquinolones for M. tuberculosis with these mutations are shown in . To determine whether there is any correlation between the MICs of the fluoroquinolones and the different mutations in the isolates, the Kruskal–Wallis test was applied. The results showed that the MIC of each drug was significantly associated with the mutations (ofloxacin: χ2=26.339, P<0.001; moxifloxacin: χ2=29.454, P<0.001; gatifloxacin: χ2=21.408, P<0.001). These results strongly suggest that the gyrA mutations are associated with the different MICs of fluoroquinolones. To further explore this association, we applied pair-wise comparisons among these four mutations using the Mann–Whitney U test. We noticed that the D94G and D94N mutations were associated with significantly higher MICs of all three drugs than were the A90V and D94A mutations ( and ).
Figure 1 MICs of the three fluoroquinolones for isolates carrying the four most frequent mutations. The transverse lines represent the median MIC for each drug. The D94G and D94N mutations were significantly associated with higher resistance to all three drugs than the A90V and D94A mutations (pair-wise Mann–Whitney U test, ).
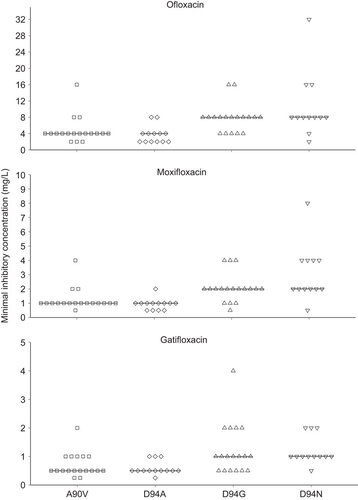
Table 2 Pair-wise comparison of resistance levels of isolates carrying A90V/D94A/D94G/D94N mutations, using the Mann–Whitney U test.
DISCUSSION
In this study, we present the phenotypic resistance profiles of 80 M. tuberculosis isolates and confirmed cross-resistance among different generations of fluoroquinolones. The obtained profiles indicated that the bactericidal activities of moxifloxacin and gatifloxacin are greater than the bactericidal activity of ofloxacin. The gyrA mutations D94G and D94N were associated with higher MICs than were the A90V and D94A mutations.
Resistance to fluoroquinolones in M. tuberculosis has been predominantly attributed to gyrA mutations.Citation10,Citation12 In this study, we found high mutation frequencies in codons 90 (18/80, 22.5%) and 94 (46/80, 57.5%). It was striking that for three different drugs, isolates with the A90V and D94A mutations showed a lower level of resistance, whereas isolates carrying the D94G and D94N mutations showed a higher level of resistance. Previous studies have indicated that fluoroquinolones inhibit the activity of the DNA gyrase complex by binding to the QBP; the binding process is shape-dependent, and mutations in the QRDRs modify the geometry of the QBP.Citation8,Citation9 We speculate that the higher resistance of M. tuberculosis carrying the D94G and D94N mutations relative to M. tuberculosis carrying the other two mutations is a result of larger QBP geometric changes, leading to greater instability. However, we noticed that this correlation was not absolute; several isolates with mutations in A90V and D94A have an extremely high level of resistance to fluoroquinolones (MIC≥16 mg/L). This phenomenon might be associated with additional resistance mechanisms, such as efflux pumps,Citation24 or the genetic background of these isolates.Citation25
Considering the limited number of effective drugs for TB treatment, identifying the resistance levels of individual mutations in M. tuberculosis is imperative to ensure that strains with a low-level of resistance are effectively treated with high-dose antibiotics or antibiotics with high bactericidal activity.Citation26 Fluoroquinolones are important second-line drugs for treating multidrug-resistant TB.Citation1 Although broad-cross resistance within fluoroquinolones has been identified, a recent study showed that the MIC of moxifloxacin was ≥4.0 mg/L, which is above moxifloxacin’s peak serum concentration and indicates possible treatment failure, for only 4% of ofloxacin-resistant isolates.Citation15 However, in this study, we found that the MIC of moxifloxacin was ≥4.0 mg/L for 11.25% (9/80) of the isolates ( and ).Citation27 Eight isolates carried mutations in D94G or D94N, which were associated with higher resistance to fluoroquinolones; these eight isolates accounted for as many as 25% of all 32 isolates carrying these mutations. Therefore, caution must be taken when using moxifloxacin to treat ofloxacin-resistant cases.
The mutations in gyrB (N498Q, A543V and G551R) were located outside the QRDR (codons 500 to 540) and have not been reported to be associated with resistance in earlier studies.Citation13,Citation14 However, in the nine isolates with gyrB mutations, six also carried additional gyrA mutations. Thus, these three gyrB mutations may not be implicated in resistance. In fact, the G551R mutation has also been detected in susceptible isolates;Citation18 therefore, we speculate that G551R may arise from a single-nucleotide polymorphism rather than being a resistance-related mutation.
In conclusion, two common mutations of gyrA, D94G and D94N, were significantly associated with higher resistance to fluoroquinolones. The MIC of moxifloxacin was ≥4.0 mg/L for a considerable proportion of clinical isolates carrying these two mutations, which might lead to treatment failure. Understanding the relationship between MICs and mutations in ofloxacin-resistant TB cases will be helpful when new-generation fluoroquinolones are used for TB treatment.
This work was supported by grants from the Key Project of the Chinese National Programs, China (2013ZX10004903-006) and the National Natural Science Foundation of China (31301033).
- Falzon D, Jaramillo E, Schünemann HJ et al.WHO guidelines for the programmatic management of drug-resistant tuberculosis: 2011 update. Eur Respir J2011;38: 516–528.
- Maruri F, Sterling TR, Kaiga AW et al.A systematic review of gyrase mutations associated with fluoroquinolone-resistant Mycobacterium tuberculosis and a proposed gyrase numbering system. J Antimicrob Chemother2012;67: 819–831.
- World Health Organization. Multidrug and extensively drug-resistant TB (M/XDR-TB): 2010 global report on surveillance and response.Geneva: WHO, 2010.Available at http://www.who.int/tb/features_archive/m_xdrtb_facts/en/ (accessed 20 December 2013).
- Aubry A, Pan XS, Fisher LM et al.Mycobacterium tuberculosis DNA gyrase: interaction with quinolones and correlation with antimycobacterial drug activity. Antimicrob Agents Chemother2004;48: 1281–1288.
- Xu C, Kreiswirth BN, Sreevatsan S et al.Fluoroquinolone resistance associated with specific gyrase mutations in clinical isolates of multidrug-resistant Mycobacterium tuberculosis. J Infect Dis1996;174: 1127–1130.
- Cabral JHM, Jackson AP, Smith CV et al.Crystal structure of the breakage–reunion domain of DNA gyrase. Nature1997;388: 903–906.
- Piton J, Petrella S, Delarue M et al.Structural insights into the quinolone resistance mechanism of Mycobacterium tuberculosis DNA gyrase. PLoS ONE2010;5: e12245.
- Tretter EM, Schoeffler AJ, Weisfield SR et al.Crystal structure of the DNA gyrase GyrA N-terminal domain from Mycobacterium tuberculosis. Proteins2010;78: 492–495.
- Aubry A, Veziris N, Cambau E et al.Novel gyrase mutations in quinolone-resistant and -hypersusceptible clinical isolates of Mycobacterium tuberculosis: functional analysis of mutant enzymes. Antimicrob Agents Chemother2006;50: 104–112.
- von Groll A, Martin A, Jureen P et al.Fluoroquinolone resistance in Mycobacterium tuberculosis and mutations in gyrA and gyrB. Antimicrob Agents Chemother2009;53: 4498–4500.
- Yin X, Yu Z.Mutation characterization of gyrA and gyrB genes in levofloxacin-resistant Mycobacterium tuberculosis clinical isolates from Guangdong Province in China. J Infect2010;61: 150.
- Sirgel FA, Warren RM, Streicher EM et al.gyrA mutations and phenotypic susceptibility levels to ofloxacin and moxifloxacin in clinical isolates of Mycobacterium tuberculosis. J Antimicrob Chemother2012;67: 1088–1093.
- Malik S, Willby M, Sikes D et al.New insights into fluoroquinolone resistance in Mycobacterium tuberculosis: functional genetic analysis of gyrA and gyrB mutations. PLoS ONE2012;7: e39754.
- Pantel A, Petrella S, Veziris N et al.Extending the definition of the GyrB quinolone resistance-determining region in Mycobacterium tuberculosis DNA gyrase for assessing fluoroquinolone resistance in M. tuberculosis. Antimicrob Agents Chemother2012;56: 1990–1996.
- Kam KM, Yip CW, Cheung TL et al.Stepwise decrease in moxifloxacin susceptibility amongst clinical isolates of multidrug-resistant Mycobacterium tuberculosis: correlation with ofloxacin susceptibility. Microb Drug Resist2006;12: 7–11.
- Rustomjee R, Lienhardt C, Kanyok T et al.A Phase II study of the sterilising activities of ofloxacin, gatifloxacin and moxifloxacin in pulmonary tuberculosis. Int J Tuberc Lung Dis2008;12: 128–138.
- Ginsburg AS, Grosset JH, Bishai WR.Fluoroquinolones, tuberculosis, and resistance. Lancet Infect Dis2003;3: 432–442.
- Cui Z, Wang J, Lu J et al.Association of mutation patterns in gyrA/B genes and ofloxacin resistance levels in Mycobacterium tuberculosis isolates from East China in 2009. BMC Infect Dis2011;11: 78.
- McNerney R, Maeurer M, Abubakar I et al.Tuberculosis diagnostics and biomarkers: needs, challenges, recent advances, and opportunities. J Infect Dis2012;205 Suppl 2: S147–S158.
- Magdalena J, Vachée A, Supply P et al.Identification of a new DNA region specific for members of Mycobacterium tuberculosis complex. J Clin Microbiol1998;36: 937–943.
- Moore DA, Evans CA, Gilman RH et al.Microscopic-observation drug-susceptibility assay for the diagnosis of TB. N Engl J Med2006;355: 1539–1550.
- Devasia RA, Blackman A, May C et al.Fluoroquinolone resistance in Mycobacterium tuberculosis: an assessment of MGIT 960, MODS and nitrate reductase assay and fluoroquinolone cross-resistance. J Antimicrob Chemother2009;63: 1173–1178.
- World Health Organization. Policy guidance on drug-susceptibility testing (DST) of second-line antituberculosis drugs.Geneva: WHO, 2008.Available at http://www.who.int/tb/publications/2008/whohtmtb_2008_392/en/index.html (accessed 20 December 2013).
- Da SPE, von Groll A, Martin A et al.Efflux as a mechanism for drug resistance in Mycobacterium tuberculosis. FEMS Immunol Med Microbiol2011;63: 1–9.
- van der Heijden YF, Maruri F, Blackman A et al.Fluoroquinolone susceptibility in Mycobacterium tuberculosis after pre-diagnosis exposure to older- versus newer-generation fluoroquinolones. Int J Antimicrob Agents2013;42: 232–237.
- World Health Organization. Stop TB Dept.Treatment of tuberculosis: guidelines.Geneva: WHO, 2010.Available at https://extranet.who.int/iris/restricted/handle/10665/44165?locale=en.
- Alffenaar JW, van Altena R, Bokkerink HJ et al.Pharmacokinetics of moxifloxacin in cerebrospinal fluid and plasma in patients with tuberculous meningitis. Clin Infect Dis2009;49: 1080–1082.
