Abstract
Bacteria belonging to the genus Cronobacter spp. have been recognized as causative agents of life-threatening systemic infections, primarily in premature, low-birth weight and/or immune-compromised neonates. Knowledge remains scarce regarding the underlying molecular mechanisms of disease development. In this study, we evaluated the use of a zebrafish model to study the pathogenesis of Cronobacter turicensis LMG 23827T, a clinical isolate responsible for two fatal sepsis cases in neonates. Here, the microinjection of approximately 50 colony forming units (CFUs) into the yolk sac resulted in the rapid multiplication of bacteria and dissemination into the blood stream at 24 h post infection (hpi), followed by the development of a severe bacteremia and larval death within 3 days. In contrast, the innate immune response of the embryos was sufficiently developed to control infection after the intravenous injection of up to 104 CFUs of bacteria. Infection studies using an isogenic mutant devoid of surviving and replicating in human macrophages (ΔfkpA) showed that this strain was highly attenuated in its ability to kill the larvae. In addition, the suitability of the zebrafish model system to study the effectiveness of antibiotics to treat Cronobacter infections in zebrafish embryos was examined. Our data indicate that the zebrafish model represents an excellent vertebrate model to study virulence-related aspects of this opportunistic pathogen in vivo.
Introduction
Cronobacter spp. are regarded as opportunistic facultative intracellular pathogens associated with the ingestion of contaminated reconstituted infant formula and cause serious illness predominantly in low-birth-weight preterm and neonatal infants.Citation1,Citation2 The clinical presentation of Cronobacter infections include necrotizing enterocolitis (NEC), bacteremia and meningitis, with case fatality rates ranging between 40% and 80%.Citation3,Citation4
The genus Cronobacter spp. - as proposed in 2008 - currently consists of seven species according to the “List of prokaryotic names with standing in nomenclature” and encompasses organisms that have previously been identified as Enterobacter sakazakii.Citation5,Citation6,Citation7 Recently, the extension of the genus Cronobacter by three more Enterobacter species was proposed by Brady et al.Citation8; however, re-examination of the biological basis for this suggestion as performed in the study by Stephan et al.Citation9 does not support further revision of the taxon at this time.
Epidemiological studies and in vitro mammalian tissue culture assays have shown that Cronobacter isolates demonstrate a variable virulence phenotype. To date, only isolates of C. sakazakii, C. malonaticus and C. turicensis have been linked to infantile infections.Citation10
Despite the progressive increase in research on Cronobacter pathogenesis in the last decade, knowledge of the exact mechanisms of infection remains fragmentary.Citation11
As an orally transmitted pathogen, Cronobacter is thought to gain entrance into the human body through the gastrointestinal tract, where it may cause NEC or, by unknown mechanism(s), may enter the systemic circulation without the manifestation of NEC.Citation12 Once the bacteria have entered the blood stream, they exhibit a tropism towards the central nervous system, showing an increased propensity to cause meningitis among low-birth-weight neonates and infants, while causing bacteremia or sepsis among slightly higher birth-weight infants.Citation13 After crossing the blood brain barrier, the pathogen enters the brain, where it causes ventriculitis, which can lead to the development of hydrocephalus, or forms other sequelae, such as cysts or brain abscesses.Citation3,Citation14
Most of the data available today have been acquired from in vitro studies. In vivo studies to confirm and extend these observations from cell culture have largely been concentrated on the neonatal rat, mouse or gerbil as model organism.Citation15,Citation16,Citation17,Citation18 Although valuable information has been obtained from these studies, the lack of possibilities for a real-time analysis and the need for laborious and invasive sample analysis limit the use of mammalian experimental animals.
The nematode Caenorhabditis elegans has been alternatively used to study Cronobacter virulence factors, exploiting the amenities of the nematode system, such as easy cultivation and transparency.Citation19 However, invertebrates are genetically not closely related to humans, and their immune system shows many differences from the human immune system. Hence, other models are needed to address specific questions related to the innate immune response to a specific pathogen in detail.
The zebrafish (Danio rerio) may be considered a hybrid between the mouse and invertebrate infection models. The most impressive feature of this model is the possibility of performing non-invasive, high-resolution, long-term time-lapse and time-course experiments to visualize infection dynamics with fluorescent markers in the transparent embryo. The small size, ease of breeding, high fertility and genetic tractability of the zebrafish are further favorable features that make the zebrafish embryo an attractive model organism. Furthermore, the zebrafish immune system displays many similarities to that of mammals, with counterparts for most of the human immune cell types.Citation20 The zebrafish innate immune system starts to develop as early as 24 h post fertilization (hpf) with primitive macrophages followed by neutrophils at 32-48 hpf. The development of the adaptive immune system lags,Citation21 which provides an opportunity to study independently the innate immunity of the larvae during the first days post fertilization (dpf). This situation sets zebrafish apart from both in vitro and mammalian in vivo infection models. Zebrafish larvae have previously been used to study infections of other bacterial pathogens,Citation22 including Listeria monocytogenes, Salmonella Typhimurium and Shigella flexneri.Citation23,Citation24,Citation25
In this study, we exploited the advantages of the zebrafish to investigate infections by Cronobacter turicensis LMG 23827T in vivo. We show here that Cronobacter causes lethal infection in zebrafish larvae, with similarities to human cases. After having successfully established the experimental parameters, the model was evaluated using a strain devoid of a gene that has recently been described as a virulence factor in C. turicensis. In addition, the model was used to study the effectiveness of different antibiotics to treat Cronobacter infection.
MATERIALS AND METHODS
Bacterial strains and growth conditions
The bacterial strains used in this study are listed in . C. turicensis LMG 23827T, a clinical isolate responsible for the death of two neonates in Zurich in 2006 has been object of previous research.Citation26,Citation29,Citation30,Citation31, Construction of the C. turicensis LMG 23827T ΔfkpA mutant, the complemented mutant C. turicensis LMG 23827T ΔfkpA::fkpATetR, as well as the mutant carrying the complementation vector pCCR9TetR only (C. turicensis LMG 23827T ΔfkpA::pCCR9TetR) has been described in detail in the study by Eshwar et al.Citation28
Table 1 Bacterial strains and mutants used in this study
The green fluorescent protein (GFP)-expressing strain C. turicensis LMG 23827T::GFPKanR was constructed by Schmid et al.Citation27 For selection purposes, during zebrafish embryo infection experiments, C. turicensis LMG 23827T::dsREDTetR, C. turicensis LMG 23827T ΔfkpA::dsREDTetR as well as E. coli DH5α::dsREDTetR were constructed in this study by transformation of vector pRZT3::dsREDTetR using standard methods. Plasmid pRZT3::dsREDTetR carrying the red fluorescent protein was a kind gift by A. M. van der Sar (VU University Medical Center, Department of Medical Microbiology and Infection Control, Netherlands).
For cultivation, the strains were grown in 10 mL of Luria–Bertani (LB, Difco, Beckton, Dickinson and Company, Allschwil, Switzerland) broth overnight at 37 °C with gentle shaking. C. turicensis LMG 23827T variants/mutants were cultivated in LB broth supplemented where appropriate with either tetracycline at 50 mg/L or kanamycin at 50 mg/L.
For microinjection experiments, the bacteria were harvested by centrifugation at 5000g for 10 min and washed once in 10 mL of Dubelccòs phosphate buffered saline (DPBS, Life Technologies, Zug, Switzerland. After a second centrifugation step, the cells were resuspended in DPBS, and appropriate dilutions were prepared in DPBS.
Zebrafish lines and husbandry
Zebrafish (Danio rerio) strains used in this study were predominantly albino lines as well as transgenic fish of the Tg(lyz:DsRED2)nz50 line that produce red fluorescent protein in neutrophils, received as a kind gift from Professor Philip Crosier, University of Auckland (New Zealand).Citation32 Adult fish were kept at a 14/10-h light/dark cycle at a pH of 7.5 and 27 °C. Eggs were obtained from natural spawning between adult fish which were set up pairwise in separate breeding tanks. Embryos were raised in petri dishes containing E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4) supplemented with 0.3 mg/L of methylene blue at 28 °C. From 24 hpf, 0.003 % 1-phenyl-2-thiourea (PTU, Sigma-Aldrich, Buchs, Switzerland) was added to prevent melanin synthesis. As albino lines lack melanized chromophores, no PTU treatment was performed on these lines. Embryo staging was performed according to Kimmel et al.Citation33
Research was conducted with approval (NO 216/2012) from the Veterinary Office, Public Health Department, Canton of Zurich (Switzerland).
Microinjection experiments
Injections were conducted using borosilicate glass microcapillary injection needles (1 mm outside diameter×0.78 mm inside diameter, Science Products GmbH, Hofheim, Germany) and a PV830 Pneumatic PicoPump (World Precision Instruments, Sarasota, Florida, USA). Prior to injection, embryos of 2 dpf were manually dechorionated and anesthetized with 200 mg/L buffered tricaine (Sigma-Aldrich, Buchs, Switzerland). Afterwards, the embryos were aligned on an agar plate and injected with 50 to 104 colony forming units (CFUs) in a 1-2-nL volume of a bacterial suspension in DPBS either directly into the blood circulation, the hindbrain ventricle or into the yolk sac. The volume of the injected suspension was previously adjusted by injection of a droplet into mineral oil and measurement of its approximate diameter over a scale bar. To determine the actual number of CFU injected, we initially injected inoculum directly onto the agar plates; however, a more precise determination of the injected CFU can be obtained by plating five embryos separately immediately after microinjection (0 hpi).
After injection, the infected embryos were allowed to recover in a petri dish with fresh E3 medium for 15 min. To follow the infection kinetics, the embryos were transferred to 6-well plates in groups of approximately 15 embryos in 4 mL of E3 medium per well, incubated at 28 °C and observed for signs of disease and survival under a stereomicroscope twice a day.
Five embryos or larvae were collected at each time point, generally 0, 15, 24, and 48 hpi, and independently treated for bacterial enumeration. The sampled larvae were euthanized with an overdose of 4 g/L buffered tricaine and transferred to different buffers and fixatives for subsequent analyses.
Bacterial enumeration by plate counting
The larvae were transferred to 1.5-mL microfuge tubes and disintegrated by repeated pipetting and vortexing for 3 min in 1 mL of PBS supplemented with 1 % Triton X- 100 (Sigma-Aldrich, Buchs, Switzerland). Subsequently, 100 μL of this mixture was plated onto LB selective plates (i.e., tetracycline 50 mg/L for strains harboring pCCR9 or pRZT3::dsRED or kanamycin 50 mg/L for selection for C. turicensis LMG 23827T::GFP). The plates were incubated up to 48 h at 37 °C.
For survival assays, the embryos were similarly microinjected and maintained individually in 24-well plates in E3 medium at 28 °C. At regular time points after infection, the number of dead larvae was determined visually based on the absence of a heartbeat.
Drug testing
Uninfected embryos were tested for antibiotic toxicity prior to the start of this experiment by incubating embryos in E3 medium supplemented with one of three drugs. For drug screening, the infected embryos were transferred into 24-well plates after yolk injections with one embryo in each well in 1 mL of E3 medium containing either 8 mg/L ampicillin, 8 mg/L tetracycline, 4 mg/L nalidixic acid or were left untreated (no drug added). As an additional control, a set of uninfected embryos was incubated in E3 medium to determine embryo quality. A negative control group was injected with DPBS. Drugs were added to the water at the required concentrations. Samples were collected and analyzed as described above.
Statistical analysis
Statistics and graph design were performed using GraphPad Prism 6 (GraphPad Software, San Diego, United States). Experiments were performed at least three times, unless stated otherwise. The CFU of groups of individual larva at various time points and under various conditions were tested for significant differences by one-way ANOVA with Bonferroni’s post-test.
Light microscopy, fluorescence imaging and image analysis
For histological examination, whole zebrafish larvae were fixed in 4% paraformaldehyde at 4 °C and embedded in cubes of cooked egg white to position them correctly for histological sections. These cubes containing the larvae were dehydrated in an alcohol series of ascending concentrations ending in xylene and afterwards embedded in paraffin. Paraffin blocks were cut in 2-3-µm thin sections, mounted on glass slides and stained using a routine protocol with hematoxylin and eosin. Histological imaging was performed with a Leica DM LS S-203675 (Leica microsystems, Heerbrugg, Switzerland) upright light microscope.
Overview images of whole larvae were obtained with an Olympus BX61 upright light microscope with both bright field and fluorescence modules. The fluorescence filter cube used was optimized for DAPI/FITC/TRIC. For higher resolution images, 3D-image stacks of whole mount samples were prepared using a confocal laser-scanning microscope (CLSM, Leica TCS SP5, Leica Microsystems, Heerbrugg, Switzerland). GFP, dsRED and DAPI were sequentially excited with the 405 nm, 488 nm and 561 nm laser lines, respectively, with emission signals collected within the respective range of wave lengths. 3D image stacks were collected sequentially (to prevent blue-green–red channel cross-talk) according to Nyquist criteria and deconvolved using HuygensPro via the Huygens Remote Manager v2.1.2 (SVI, Netherlands). Images were further analyzed with Imaris 7.6.1 (Bitplane, Zurich, Switzerland) and aligned with Adobe Photoshop Elements 12.
Transmission electron microscopy
For transmission electron microscopy (TEM), the larvae were fixed in a mixed solution of 1 % paraformaldehyde (Sigma-Aldrich, Buchs, Switzerland) and 2.5 % glutaraldehyde (Sigma-Aldrich, Buchs, Switzerland) in 0.1 M sodium phosphate buffer, pH 7.5 at 4 °C overnight. Afterwards, the samples were prepared for embedding into epoxy resin and for transmission electron microscopy according to standard procedures. Pospischil et al., 1990). Epoxy resin blocks were screened for larvae using semithin sections (1 µm), which were stained with toluidine blue (Sigma-Aldrich, Buchs, Switzerland) to visualize tissue. Ultrathin sections (80 nm) were mounted on copper grids (Merck Eurolab AG, Dietlikon, Switzerland), contrasted with uranyl acetate dihydrate (Sigma-Aldrich, Buchs, Switzerland) and lead citrate (Merck Eurolab AG, Dietlikon, Switzerland) and investigated using a Philips CM10 transmission electron microscope (Philips Electron Optics, Eindhoven, The Netherlands). Images were processed with Imaris (Bitplane AG, Zurich, Switzerland) and assembled for publication using Adobe Photoshop.
Confocal live imaging
To perform high-resolution confocal live imaging, the injected larvae were positioned in 35-mm glass-bottom dishes (Iwaki, Eurodyne Limited, Lindale, UK). The entire larva was covered and immobilized with 1 % low-melting-point agarose solution. A total of 2 mL of fish water containing tricaine was added to cover the immobilized larvae. Confocal microscopy was performed at 26 °C. A Leica SP8 (Leica Microsystems, Heerbrugg, Switzerland) automated upright confocal laser scanning microscope allowing simultaneous acquisition of three fluorescent channels and one bright field or differential interference contrast (DIC) was used. The detection system in this microscope is equipped with two photomultiplier tubes and a hybrid detector and a 20×water immersion objective (HC PL APO NA-0.5 WD-3.5 mm) was used to image live infected larvae. The 4D images produced by the time-lapse acquisitions were processed, clipped, examined and interpreted using the Imaris software. Maximum intensity projection was used to project developed Z-stacks and files were exported in AVI format. To mount figures, frames captured from AVI files were handled using Photoshop software. Imaris software was used to crop and annotate the exported AVI files, then compressed and converted into QuickTime movies with QuickTime Pro software.
RESULTS
Infection of zebrafish larvae with Cronobacter via microinjection is lethal
To develop a Cronobacter turicensis infection model in a genetically tractable vertebrate model host, we investigated whether the strain C. turicensis LMG 23827T, a strain originally isolated from a fatal neonatal infection could lethally infect zebrafish embryos at 2 dpf. Initial experiments to infect zebrafish with this strain by immersing dechorionated embryos in a suspension of strain LMG 23827T failed. Lethality required high concentrations of strain LMG 23827T or LMG 23827T::GFP (109–1010 CFUs/mL), and the experiments were not reproducible (data not shown). Similar results were reported for co-infection experiments using other pathogens under static immersion conditions.Citation34 Moreover, using the bath immersion experimental design, we were unable to establish a stable infection in the digestive system of the larvae. Similar observations were reported in the study by Levraud et al.Citation23 showing that zebrafish embryos were not susceptible to oral infection with Listeria monocytogenes.
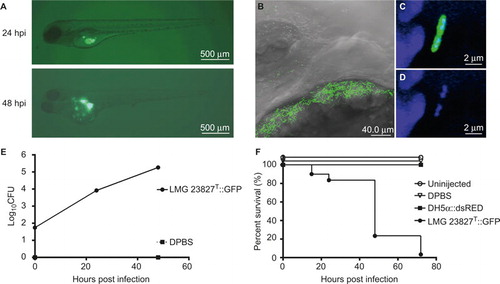
Therefore, we next focused on the possibility of introducing LMG 23827T or LMG 23827T::GFP into 2 dpf-old embryos directly by microinjection into the yolk sac, the common cardinal vein or the hindbrain ventricle, thereby exploiting the advantage of easy fluorescent tracking by using the GFP transgenic strain C. turicensis LMG 23827T::GFP with concentrations ranging from 50 to 104 CFUs. Yolk injections were performed into the posterior part of the yolk sac before the extension to prevent perforation of the common cardinal vein, which widely covers the anterior part of the yolk. After injection, the embryos were transferred into 24-well plates containing fresh E3 medium and further incubated at 28 °C. Signs of disease, larval survival, fluorescence pattern of bacteria, and bacterial load were examined over time. The bacterial load was determined by counting the CFU of homogenates of whole individual larva plated on agar plates containing kanamycin for selection purposes at 37 °C overnight.
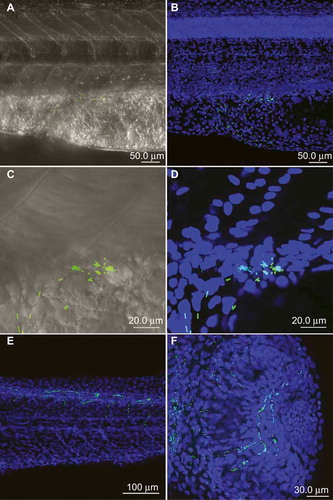
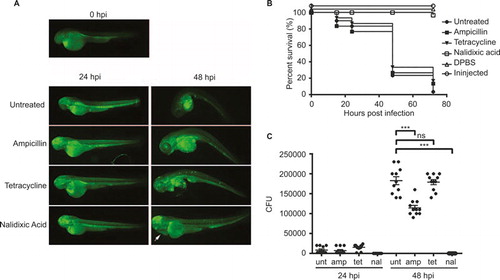
Intravenous injections of up to 104 CFUs and hindbrain injections of up to 103 CFUs did not result in an apparent infection. Bacteria were cleared from the system within the first 24 hpi, and the larvae showed no indication of disease or mortality (data not shown). However, injections of as little as 50 CFUs into the yolk resulted in the rapid replication of Cronobacter inside the yolk sac of approximately two log units within 24 hpi and a subsequent spreading into the larval blood stream between 24 and 48 hpi (). Confocal imaging revealed bacteria accumulating at the surface of the yolk sac before further spreading into the larvae (). The bacteria inside the yolk could be found dividing by binary fission (). The bacterial load continued to increase for another log unit at the same time (). Mortality after yolk injections increased up to 100 % at 72 hpi, while microinjection of equal or greater numbers of E. coli DH5α::dsRED resulted in complete survival of the infected larvae ().
The traverse of Cronobacter to the bloodstream could be observed throughout the entire length of the boundary between yolk and vasculature (), followed by an accumulation of bacterial clusters in the capillaries of the trunk and the eyes ( and ).
Infection progression and pathology
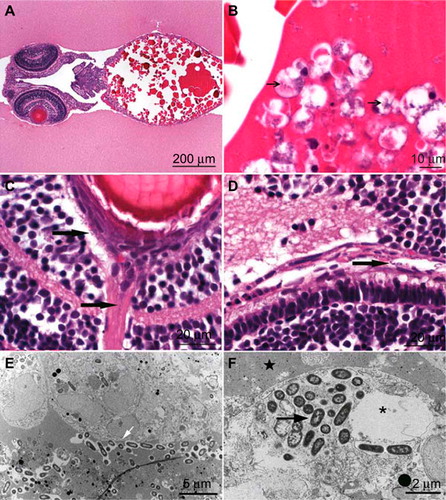
Larvae that were injected into the yolk sac showed at 30-48 hpi small 1×2 µm, rod-shaped bacteria free and intracytoplasmic in leukocytes as macrophages or neutrophils in the yolk sac () and in the lumina of several blood vessels, especially close to the eyes and brain (). The number of macrophages and neutrophils within the yolk sac lining the wall was increased compared with control animals (data not shown).
TEM images showed long rods of 2-µm length and 1-µm width that were lying free in the protein of the yolk sac (). The bacteria contained a thin cell wall, typical for gram-negative rods, and a loose chromatin pattern. Some bacteria were dividing, which could also be observed by confocal microscopy. Some leukocytes containing bacteria showed degeneration as large pale intracytoplasmic vacuolation, crystolysis and swelling of mitochondria, increase of lipid globules or dilation or even fragmentation of Golgi or endoplasmic reticulum (). Additional signs of degeneration, such as karyorhexis, karyopyknosis and hypereosinophilia of the cytoplasm, were observed in many leukocytes (). The presence of neutrophils inside the yolk sack was confirmed by confocal imaging, where labeled neutrophils were associated with the bacteria ().
Innate immune response to Cronobacter infection (CLSM Leica SP8 – live imaging)
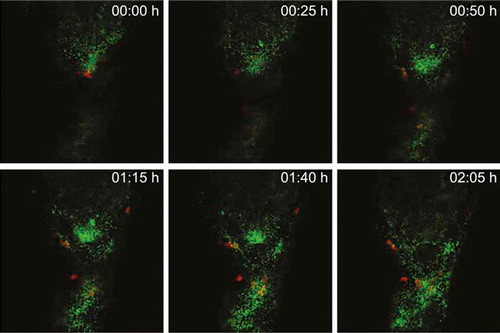
To visualize the dynamics of Cronobacter replication and the innate immune reaction to the infection inside the yolk, we utilized transgenic 2 dpf zebrafish embryos of the Tg(lyz:DsRED2)nz50 line, which harbor red fluorescent protein-expressing neutrophils. These embryos were injected with a dose of approximately 50 CFUs of GFP-expressing C. turicensis LMG 23827T::GFP into the yolk sac at 2 dpf. The Cronobacter-host interactions were captured in real-time at 24 hpi using high-resolution confocal laser scanning microscopy for a time course of approximately 2 h. We observed rapid replication of Cronobacter inside the yolk forming several clusters of motile bacteria. Over time, an increasing number of red fluorescent neutrophils was recruited to the yolk taking up the bacteria but were unable to control the proliferation and spreading of Cronobacter (, Supplementary Video).
Drug screening
For the next step, we determined whether the zebrafish model may be suitable for testing the effectiveness of antimicrobial agents to clear infections with C. turicensis LMG 23827T. Prior to these experiments, the minimum inhibitory concentrations (MICs) of a selection of antimicrobial drugs belonging to different antibiotic classes were determined for the wild type C. turicensis LMG 23827T as well as the GFP-expressing strain C. turicensis LMG 23827T::GFPKanR using E-test strips (bioMerieux, Marcy-l-Etoile, France) on Müller Hinton agar, according to the recommendations of the manufacturer. The following MIC values (in mg/L) were determined for both strains: ampicillin, 0.75; tetracycline, 1.5; cephalothin, 6; rifampicin, 2; gentamicin, 0.38; polymyxin B, 0.094; nalidixic acid, 0.5; and chloramphenicol, 64. Conversion of the MIC data into qualitative categories using the European Committee on Antimicrobial Susceptibility Testing breakpoints suggested that C. turicensis LMG 23827T was susceptible to all tested antibiotics with exception of rifampicin and chloramphenicol. There was no variation among the wild type and its GFP variant (data not shown).
Based on these findings, we tested the activity of ampicillin, tetracycline and nalidixic acid against Cronobacter in vivo in the zebrafish infection model. The embryos were tested for antibiotic toxicity prior to the start of this experiment. Embryos in E3 medium supplemented with tetracycline or ampicillin did not exhibit any toxic effect and/or mortality; however, the embryos exposed to nalidixic acid exhibited pericardial edema but no mortality (data not shown). Because all three antibiotics are water soluble, they were administered to the fish water after the yolk injections of Cronobacter or DPBS as control. Another control group was infected but left without treatment. The distribution of Cronobacter within the infected larvae was followed by fluorescence microscopy after the injection of the GFP-expressing strain (). The survival rate () and the bacterial load () were determined for individual larvae by microscopic observation and plate count enumeration. While treatment with tetracycline had no significant effect on the bacterial load and survival, treatment with ampicillin significantly reduced the bacterial load compared with untreated larvae, but did not increase survival. However, treatment with nalidixic acid had a significant impact on both bacterial load and the survival of infected larvae. At 24 hpi, Cronobacter could no longer be detected in these larvae by plate count or by fluorescence microscopy. Furthermore, the survival rate was close to 100 % at 72 hpi. Interestingly, during treatment with nalidixic acid, the formation of pericardial edema was observed in nearly 100% of all larvae.
FkpA is an important virulence factor for Cronobacter turicensis infection in zebrafish embryos
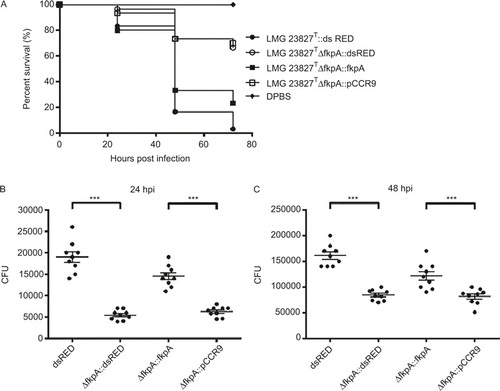
Given the previous results and observations concerning the behavior of Cronobacter and innate immune cells after injection into the yolk sac, we concluded that internalization and survival of Cronobacter cells in professional phagocytes of the innate immune system, such as macrophages present in the yolk and/or the blood stream, play a key role during the infection process. In a recent study, the eminent role of a functional FkpA (also known as macrophage infectivity potentiator-like protein) in survival and replication in human macrophages was reported for C. turicensis LMG 23827T.Citation28 We therefore tested a ΔfkpA in-frame mutant for attenuated pathogenicity in infection experiments using the above-described experimental design; C. turicensis LMG 23827T::dsRED served as control. The survival rate at 48 hpi increased to approximately 70 % for ΔfkpA::dsRED mutant-injected larvae (), thus confirming the role of this gene as a pathogenicity factor during Cronobacter infection in zebrafish embryos. Furthermore, the bacterial load in ΔfkpA::dsRED mutant (C. turicensis LMG 23827T ΔfkpA::dsRED)-injected larvae was significantly lower compared with the control group (). Injection experiments using the complemented mutant strain (C. turicensis LMG 23827T ΔfkpA::fkpA) resulted in a lower survival rate and a higher bacterial load, whereas control experiments using the mutant strain transformed with the vector alone (C. turicensis LMG 23827T ΔfkpA::pCCR9) yielded survival rates and bacterial loads comparable to the ones observed in the ΔfkpA::dsRED mutant-injection experiments ().
DISCUSSION
To adopt the zebrafish model to study these human opportunistic pathogens, several experimental designs were used. As infecting the embryos via the oral route proved unsuccessful, we focused on introducing the pathogens into the animal circulation system via microinjection. However, injection of up to 10 000 CFUs into the cardinal vein of 2 dpf old zebrafish larvae did not result in clinical signs of infection in the embryos. Fluorescence microscopy showed that no replication occurred and the bacteria were cleared instead within 24 hpi (data not shown). In fact, it has been reported that Cronobacter spp. exhibit only a “moderate” capability to survive in human blood or serum. Thus, it was shown for C. sakazakii, which is closely related to C. turicensis, that a minimum of a two-log reduction of a bacterial inoculum occurred within a 30-min exposure in 50% human blood.Citation35
In a recent study, similar observations were reported for zebrafish larvae infection studies on Staphylococcus epidermidis when comparing the results from yolk injection versus blood stream (caudal vein) injection experiments.Citation36 Within this study, it was speculated that the better survival rate of the bacteria injected in the yolk may be explained by several factors (or a combination of those), such as repeated cycles of invasion from the yolk and/or that internalization and proliferation of bacteria in the yolk results in “priming” to an infectious growth strategy but also alternations of the host immune system due to the prolonged exposure to high numbers of bacteria were suggested.
Thus, we concluded that the yolk injection is a uniquely suitable infection site. We showed that, following injection of as few as 50 CFUs, the bacteria were rapidly replicating both freely in the yolk as well as internalized in the primitive macrophages and neutrophils present in the yolk sac. The affected leucocytes were not killed rapidly, and the phagocytosed bacteria did not escape from these cells. At 24 hpi, free bacteria as well as infected macrophages were lining up near the border of the yolk sac, where a traverse into the blood vessel and the surrounding tissue was observed. At later time points (30 hpi – 48 hpi), bacteria colonized the lumina of small blood vessels, especially those close to the eyes and brain. Accumulation of bacteria in the capillaries of the eyes is a typical feature for septicemia and bacteremia in fish. At these sites, the bacteria were observed to form micro-colonies in which they replicated and from which they were shed into the circulation. Thus, during infection in zebrafish embryos, a combination of extracellular and intracellular replication of Cronobacter could be observed.
In the next experiment, a selection of antimicrobial compounds was tested for their ability to cure a Cronobacter infection in the embryos by application of the drugs to the fish water. Given that the antibiotic concentrations applied were previously shown to be lethal for the bacteria in vitro, we can only speculate why the ampicillin and tetracycline treatments were not effective in vivo. One explanation may be that the molecules are not reaching lethal concentrations at the predominant infection site inside the yolk, which could be influenced by the size of the molecules, the local pH or the acid/base character of the antimicrobial substance. Nalidixic acid was the only drug to prove efficient in killing the bacteria inside the host. Although the survival rate of infected fishes was close to 100% after 72 hpi, pericardial edema was observed during drug treatment. Interestingly, similar adverse effects, namely, increased intracranial pressure leading to the formation of cerebral edema, have been reported in humans, especially in infants and young children after treatment with nalidixic acid.Citation37 This result also underlines that zebrafish larvae can show typical reactions to other influencing factors, such as antibiotic treatment, and can thus be used as a drug screening model not only to test the efficacy of a drug against the pathogen in vivo but also to predict possible adverse effects on the patient.
It has been shown that Cronobacter spp. are capable of surviving and replicating within human macrophages for up to 96 h, and, in a recent study, the FkpA protein was identified as a key factor involved in this ability. When using an fkpA mutant that is defective in survival and replication in human macrophages, pathogenicity was strongly attenuated in the zebrafish model. This finding provides evidence that macrophages and other phagocytic cells can play a crucial role in the process of traversing physical barriers such as epithelia and endothelia and, by this, promote further systemic spreading of intracellular bacterial pathogens within the host organism.
Taken together, by exploiting the favorable features of the zebrafish within this study, we have established and applied the first zebrafish model to study the pathogenesis of C. turicensis in vivo. Our model provides interesting insights into the pathogenic nature of these opportunistic facultative intracellular pathogens and also shows the reaction of the innate immune system to an infection in real time. With the experimental design developed in this study, the results can be obtained within days, and the number of experiments can be easily scaled up. Thus, large numbers of bacterial mutants and strains can be screened for virulence-related factors of Cronobacter, providing a promising tool to discover further detailed features of Cronobacter virulence and the counteracting innate immune response in future studies.
Supplementary matierial 1
Download MPEG Video (3.4 MB)This work was supported by the Swiss National Science Foundation grant 310030_138533/1. We would like to thank Lisbeth Nufer (Institute of Veterinary Pathology, Vetsuisse Faculty, University of Zurich, Zurich, Switzerland) for the TEM analysis.
Supplementary Information for this article can be found on the Emerging Microbes & Infections website (http://www.nature.com/emi)
- Bar-Oz B, Preminger A, Peleg O, Block C, Arad I.Enterobacter sakazakii infection in the newborn. Acta Paediatr 2001; 90: 356–358.
- Hunter CJ, Bean JF.Cronobacter: an emerging opportunistic pathogen associated with neonatal meningitis, sepsis and necrotizing enterocolitis. J Perinatol 2013; 33: 581–585.
- Bowen AB, Braden CR.Invasive Enterobacter sakazakii disease in infants. Emerg Infect Dis 2006; 12: 1185–1189.
- Friedemann M.Epidemiology of invasive neonatal Cronobacter (Enterobacter sakazakii) infections. Eur J Clin Microbiol Infect Dis 2009; 28: 1297–1304.
- Euzéby JP. List of Prokaryotic names with Standing in Nomenclature.Paris: LPSN, 1997.Available at http://www.bacterio.net (accessed 12 March 2015).
- Iversen C, Mullane N, McCardell B et al.Cronobacter gen. nov., a new genus to accommodate the biogroups of Enterobacter sakazakii, and proposal of Cronobacter sakazakii gen. nov., comb. nov., Cronobacter malonaticus sp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov., Cronobacter genomospecies 1, and of three subspecies, Cronobacter dublinensis subsp. dublinensis subsp. nov., Cronobacter dublinensis subsp. lausannensis subsp. nov. and Cronobacter dublinensis subsp. lactaridi subsp. nov. Int J Syst Evol Microbiol 2008; 58: 1442–1447.
- Joseph S, Cetinkaya E, Drahovska H, Levican A, Figueras MJ, Forsythe SJ.Cronobacter condimenti sp. nov., isolated from spiced meat, and Cronobacter universalis sp. nov., a species designation for Cronobacter sp. genomospecies 1, recovered from a leg infection, water and food ingredients. Int J Syst Evol Microbiol 2012; 62: 1277–1283.
- Brady C, Cleenwerck I, Venter S, Coutinho T, De Vos P.Taxonomic evaluation of the genus Enterobacter based on multilocus sequence analysis (MLSA): Proposal to reclassify E. nimipressuralis and E. amnigenus into Lelliottia gen. nov. as Lelliottia nimipressuralis comb. nov. and Lelliottia amnigena comb. nov., respectively, E. gergoviae and E. pyrinus into Pluralibacter gen. nov. as Pluralibacter gergoviae comb. nov. and Pluralibacter pyrinus comb. nov., respectively, E. cowanii, E. radicincitans, E. oryzae and E. arachidis into Kosakonia gen. nov. as Kosakonia cowanii comb. nov., Kosakonia radicincitans comb. nov., Kosakonia oryzae comb. nov. and Kosakonia arachidis comb. nov., respectively, and E. turicensis, E. helveticus and E. pulveris into Cronobacter as Cronobacter zurichensis nom. nov., Cronobacter helveticus comb. nov. and Cronobacter pulveris comb. nov., respectively, and emended description of the genera Enterobacter and Cronobacter. Syst Appl Microbiol 2013; 36: 309–319.
- Stephan R, Grim CJ, Gopinath GR et al.Re-examination of the taxonomic status of Enterobacter helveticus, Enterobacter pulveris, and Enterobacter turicensis as members of Cronobacter and description of Siccibacter turicensis com. nov., Franconibacter helveticus comb. nov., and Franconibacter pulveris com. nov. Int J Syst Evol Microbiol 2014; 64: 3402–3410.
- Joseph S, Forsythe SJ.Predominance of Cronobacter sakazakii sequence type 4 in neonatal infections. Emerg Infect Dis 2011; 17: 1713–1715.
- Jaradat ZW, Al Mousa W, Elbetieha A, Al Nabulsi A, Tall BD.Cronobacter spp.– opportunistic food-borne pathogens. A review of their virulence and environmental-adaptive traits. J Med Microbiol 2014; 63: 1023–1037.
- van Acker J, de Smet F, Muyldermans G, Bougatef A, Naessens A, Lauwers S.Outbreak of necrotizing enterocolitis associated with Enterobacter sakazakii in powdered milk formula. J Clin Microbiol 2001; 39: 293–297.
- Yan QQ, Condell O, Power K, Butler F, Tall BD, Fanning S.Cronobacter species (formerly known as Enterobacter sakazakii) in powdered infant formula: a review of our current understanding of the biology of this bacterium. J Appl Microbiol 2012; 113: 1–15.
- Chenu JW, Cox JM.Cronobacter (‘Enterobacter sakazakii’): current status and future prospects. Lett Appl Microbiol 2009; 49: 153–159.
- Townsend SM, Hurrell E, Gonzalez-Gomez I et al.Enterobacter sakazakii invades brain capillary endothelial cells, persists in human macrophages influencing cytokine secretion and induces severe brain pathology in the neonatal rat. Microbiology 2007; 153: 3538–3547.
- Mittal R, Wang Y, Hunter CJ, Gonzalez-Gomez I, Prasadarao NV.Brain damage in newborn rat model of meningitis by Enterobacter sakazakii: a role for outer membrane protein A. Lab Invest 2009; 89: 263–277.
- Pagotto FJ, Farber JM.Cronobacter spp. (Enterobacter sakazakii): Advice, policy and research in Canada. Int J Food Microbiol 2009; 136: 238–245.
- Lee HA, Hong S, Park H, Kim H, Kim O.Cronobacter sakazakii infection induced fatal clinical sequels including meningitis in neonatal ICR mice. Lab Anim Res 2011; 27: 59–62.
- Sivamaruthi BS, Ganguli A, Kumar M, Bhaviya S, Pandian SK, Balamurugan K.Caenorhabditis elegans as a model for studying Cronobacter sakazakii ATCC BAA-894 pathogenesis. J Basic Microbiol 2011; 51: 540–549.
- Meeker ND, Trede NS.Immunology and zebrafish: spawning new models of human disease. Dev Comp Immunol 2008; 32: 745–757.
- Meijer AH, Spaink HP.Host-pathogen interactions made transparent with the zebrafish model. Curr Drug Targets 2011; 12: 1000–1017.
- Kanther M, Rawls JF.Host-microbe interactions in the developing zebrafish. Curr Opin Immunol 2010; 22: 10–19.
- Levraud JP, Disson O, Kissa K et al.Real-time observation of Listeria monocytogenes-phagocyte interactions in living zebrafish larvae. Infect Immun 2009; 77: 3651–3660.
- van der Sar AM, Musters RJ, van Eeden FJ, Appelmelk BJ, Vandenbroucke-Grauls CM, Bitter W.Zebrafish embryos as a model host for the real time analysis of Salmonella Typhimurium infections. Cell Microbiol 2003; 5: 601–611.
- Mostowy S, Boucontet L, Mazon Moya MJ et al.The zebrafish as a new model for the in vivo study of Shigella flexneri interaction with phagocytes and bacterial autophagy. PLoS Pathog 2013; 9: e1003588.
- Essers B, Baenzinger O, Huisman TA et al.Neonatal sepsis with Enterobacter sakazakii in premature twins. Swiss Medical Weekly 2006; 136 (Suppl 151): 22S
- Schmid M, Iversen C, Gontia I et al.Evidence for a plant-associated natural habitat for Cronobacter spp. Res Microbiol 2009; 160: 608–614.
- Eshwar AK, Tasara T, Stephan R, Lehner A.Influence of FkpA variants on survival and replication of Cronobacter spp. in human macrophages. Res Microbiol 2015; 166: 186–195.
- Stephan R, Lehner A, Tischler P, Rattei T.Complete genome sequence of Cronobacter turicensis LMG 23827, a food-borne pathogen causing deaths in neonates. J Bacteriol 2011; 193: 309–310.
- Carranza P, Hartmann I, Lehner A et al.Proteomic profiling of Cronobacter turicensis 3032, a food-borne opportunistic pathogen. Proteomics 2009; 9: 3564–3579.
- Carranza P, Grunau A, Schneider T et al.A gel-free quantitative proteomics approach to investigate temperature adaptation of the food-borne pathogen Cronobacter turicensis 3032. Proteomics 2010; 10: 3248–3261.
- Hall C, Flores MV, Storm T, Crosier K, Crosier P.The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC Dev Biol 2007; 7: 42.
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF.Stages of embryonic development of the zebrafish. Dev Dyn 1995; 203: 253–310.
- van Soest JJ, Stockhammer OW, Ordas A, Bloemberg GV, Spaink HP, Meijer AH.Comparison of static immersion and intravenous injection systems for exposure of zebrafish embryos to the natural pathogen Edwardsiella tarda. BMC Immunol 2011; 12: 58.
- Schwizer S, Tasara T, Zurfluh K, Stephan R, Lehner A.Identification of genes involved in serum tolerance in the clinical strain Cronobacter sakazakii ES5. BMC Microbiol 2013; 13: 38.
- Veneman WJ, Stockhammer OW, de Boer L, Zaat SA, Meijer AH, Spaink HP.A zebrafish high throughput screening system used for Staphylococcus epidermidis infection marker discovery. BMC Genomics 2013; 14: 255
- Rao KG.Pseudotumor cerebri associated with nalidixic acid. Urology 1974; 4: 204–207.