Abstract
Fluoroquinolone resistance is gradually acquired through several mechanisms. In particular, chromosomal mutations in the genes encoding topoisomerases II and IV and increased expression of the multidrug efflux pump AcrAB-TolC are the most common mechanisms. In this study, multiplex polymerase chain reaction (PCR) protocols were designed for high-throughput sequencing of the quinolone resistance determining regions of topoisomerases genes (gyrA, parC and parE) and/or the expression regulation systems of multidrug efflux pump AcrAB (acrRAB, marRAB and soxSR). These protocols were applied to sequence samples from five subpopulations of 103 clinical Escherichia coli isolates. These subpopulations were classified according to their levofloxacin susceptibility pattern as follows: highly resistant (HR), resistant (R), intermediate (I), reduced susceptibility (RS) and susceptible (S). All HR isolates had mutations in the six genes surveyed, with two ‘supermutator’ isolates harboring 13 mutations in these six genes. Strong associations were observed between mutations in acrR and HR isolates, parE and R/HR isolates and parC and I/R/HR isolates, whereas surprisingly, gyrA mutations were common in RS/I/R/HR isolates. Further investigation revealed that strong associations were limited to the triple mutations gyrA-S83L/D87N/R237H and HR isolates and the double mutations S83L/D87N and I/R/HR isolates, whereas the single mutation S83L was common in RS/I/R/HR isolates. Interestingly, two novel mutations (gyrA-R237H and acrR-V29G) were located and found to strongly associate with HR isolates. To the best of our knowledge, the gyrA-R237H and acrR-V29G mutations have never been reported and require further investigation to determine their exact role in resistance or ‘fitness’ as defined by their ability to compensate for the organismal cost of gaining resistance.
Introduction
Fluoroquinolones (FQs) are bactericidal antimicrobial agents that target topoisomerase II (DNA gyrase) and topoisomerase IV enzymes. They inhibit DNA synthesis by binding to the enzyme–DNA complex and stabilizing DNA strand breaks created by the enzyme.Citation1 The increasing FQ resistance rates in many bacterial species in recent years is a major concern for the healthcare community.Citation2,Citation3 FQ resistance is acquired through chromosomal mutations, at a rate of 5×10−9–5×10−7, in genes encoding DNA gyrase (gyrA and gyrB) and topoisomerase IV (parC and parE).Citation4,Citation5,Citation6 FQ resistance can also be acquired through increased expression of the multidrug efflux pump AcrAB due to mutations in operons (acrRAB, marRAB and soxSR).Citation7,Citation8,Citation9 Other mechanisms include the acquisition of the plasmid-borne genes qnr (prevalence of <2%) and aac (6′)-Ib-cr (prevalence of 25%) that encode drug-blocking and drug-modifying proteins, respectively, in addition to other unknown mechanisms.Citation4,Citation5
Mutations in topoisomerase genes occur in the quinolone-resistance determining region (QRDR). These mutations cause the mutant enzyme–DNA complex to have a lower affinity to the drug, specifically at positions 83 or 87 for the GyrA subunit of DNA gyrase and positions 78, 80 or 84 for the ParC subunit of topoisomerase IV.Citation10,Citation11 Mutations in acrR, marR and soxR, which are the repressor genes of operons acrRAB, marRAB and soxSR, respectively, cause the overexpression of the multidrug efflux pump AcrAB and its direct activators MarA and SoxS. In addition, MarA and SoxS downregulate the outer membrane porin OmpF, resulting in decreased cell permeability and resistance to oxidants as well as antibiotics.Citation7,Citation8,Citation12,Citation13
From this perspective, the present study attempted to investigate topoisomerase mutations (in gyrA, parC and parE) and AcrAB efflux pump regulator mutations (in acrR, marRA and soxSR) in Escherichia coli and associate these mutations with our proposed different levels of levofloxacin resistance. Cavaco et al.Citation14 made similar efforts to correlate resistance mechanisms to minimum inhibitory concentrations (MICs) in quinolone-resistant E. coli isolates.
The Clinical and Laboratory Standards Institute (CLSI) publishes annual guidelines for antimicrobial susceptibility testing and the interpretation breakpoints of a large number of organism–drug combinations.Citation15 For quinolones, the CLSI provides a three-level interpretation (resistant (R), intermediate (I) and susceptible (S)) that is primarily for clinical use. In this study, the distribution of the isolates covered 16 MIC dilutions and some isolates scored MIC values ×1000 greater than wild-type MIC values. It was necessary to cluster this broad distribution into a number of manageable classes that could be correlated to the genotypes under investigation. Therefore, attempts were made to establish a five-level interpretation system using ‘epidemiological’ breakpoints that were generated by a statistical method similar to the method of Kronvall.Citation16 The resultant categories were labeled as follows: highly resistant (HR), R, I, reduced susceptibility (RS) and S. These classifications were satisfactory for the purposes of this study.
MATERIALS AND METHODS
Chemicals, biochemical reagents and media
Mueller-Hinton agar and broth were from LabM (Lancashire, UK); Luria-Bertani (LB) medium was from CONDA Pronadisa (Madrid, Spain). Levofloxacin (5 mg/mL) was purchased from Sanofi-Aventis (Cairo, Egypt); levofloxacin disks (5 µg) were from Bioanalyse (Ankara, Turkey).
Bacterial isolates
A total of 103 clinical E. coli isolates were used in this study. They were under investigation for their virulence factors in two separate, previous studies. Eighty-eight isolates were associated with enteric infections,Citation17 and 15 were associated with urogenital infections (El-Far M et al., unpublished data, 2014). Therefore, these isolates represented convenient samples that were received as identified stock cultures preserved at −80 °C in 15% glycerol. E. coli K-12 was kindly provided by the Biotechnology Centre, Faculty of Pharmacy at Cairo University.Citation18 Unless otherwise specified, all isolates were propagated aerobically at 35 °C in LB broth or on LB agar. Stock cultures were stored at −80 °C in 15% glycerol.
Antimicrobial susceptibility testing
Susceptibility profiles were initially screened using the disk diffusion method and verified by determining the MIC using the broth microdilution method. Both methods were performed according to the CLSI 2012 documents M02-A11 (for the disk diffusion method) and M07-A9 (for the broth microdilution method).Citation14 E. coli K-12 was included as a control and the same lots of broth and agar media were used for susceptibility testing of all isolates.
Instead of considering the collected isolates as a single population with a normal distribution, they were treated as a mixture of different subpopulations, each with its own normal distribution. To define the subpopulations in MIC and inhibition zone histograms, the modal MICs and zone diameters on the histograms were located and the cutoff values around the mean of each subpopulation were calculated to be at ±2.0 standard deviations (SDs). The adopted method is similar to the normalized resistance interpretation method,Citation16 which is used to establish epidemiological breakpoints separating wild-type isolate subpopulations from those harboring resistance mechanisms.
Polymerase chain reaction (PCR) amplification
Initially, the PrimerQuest program (IDT, Coralville, IA, USA) was used to design primers in the conserved regions flanking the target region of each gene, which were located by blasting the sequences from E. coli K-12 MG1655 using NCBI’s BLAST for the taxid Escherichia. Primer compatibility and specificity in the multiplex reaction were checked using OligoAnalyzer 3.1 (IDT) and Primer-BLAST software from NCBI,Citation19 respectively. The six newly designed pairs of oligonucleotide primers are listed in .
Table 1 A list of the designed primers in this study that were used for amplification and sequencing
Multiplex PCR protocols were designed for the simultaneous amplification of the QRDRs of topoisomerase genes (gyrA, parC and parE) and/or the genes involved in regulating the expression of the AcrAB efflux pump in their entirety (acrR, marRA operon and soxSR operon) (with a maximum of six PCR products). The PCRs were performed using the EmeraldAmp GT PCR Master Mix (Takara, Otsu, Shiga, Japan) in a TECHNE thermocycler (Bibby Scientific Ltd, Staffordshire, UK). Colony lysates of each bacterial isolate were used as the PCR amplification template. Multiplex PCR was optimized by varying the cycling conditions and primer ratios until the final settings listed in were reached.
Table 2 Conditions for the uniplex and multiplex amplification reactions
Gel electrophoresis and DNA extraction
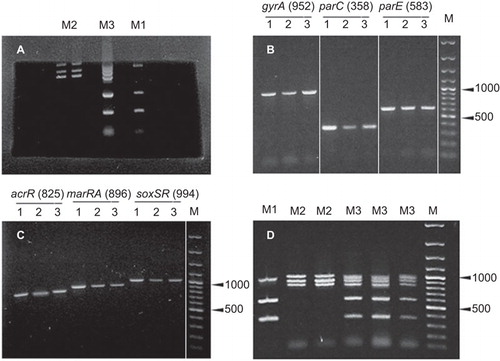
DNA fragments from the multiplex PCRs were separated using the ADVANCE electrophoresis system (Mupid-exu, Chuo-ku, Tokyo, Japan) at 5 V/cm and a combination of 1% agarose and 0.2% agarose (Lonza SeaKem LE Agarose, Walkersville, MD, USA). A similar method was described in a previous study, whereby sequencing of the separated fragments was performed directly with little additional purification.Citation20 A 1% agarose gel was prepared using a gel mold, subsequently, a rectangle was cut out from the interior portion and a 0.2% agarose gel was poured into this space (). The fragments were initially separated on the 1% agarose portion of the gel and then allowed to enter the 0.2% portion of the gel, where they were cut, pooled, and purified using a GeneJET Gel Extraction Kit (Thermo Scientific, Vilnius, Lithuania). Because of the subsequent purification and dilution steps, this method guaranteed that agarose concentrations were <0.2% in the sequencing reaction, because agarose concentrations above 0.2% could inhibit the reaction.Citation21
DNA sequencing
Mutations in the QRDRs of the gyrA, parC and parE genes and mutations in the entire genes of the marRA system, soxSR system and acrR were identified by sequencing the purified PCR amplicons using the dideoxynucleotide chain termination methodCitation22 with fluorescent cycle sequencing using dye-labeled terminators (BigDye Terminator version 3.1 cycle sequencing kit; Applied Biosystems, Grand Island, NY, USA) on an ABI prism 3730 automated DNA sequencer.
Genetic analysis
The online software Clustal Omega (EMBL-EBI, Hinxton, UK)Citation23 was used to perform the multiple sequence alignment. A mutation was considered evident if it resulted in a unique amino acid change when compared to the publicly available NCBI sequences of the E. coli K-12 susceptible substrains MG1655 and DH10B.
Statistical analysis
Data were analyzed using the Statistical Package for the Social Sciences software version 19.0 (SPSS Inc., Chicago, IL, USA). The general association of mutations with MIC values was analyzed using the Kruskal–Wallis (KW) test. The difference between isolates with and without mutations at specific MIC values was assessed using the Fisher’s exact (FE) test. A P value of <0.05 for a whole family of tests was considered statistically significant. The Bonferroni correction method was used to re-correct for multiple comparisons in the KW and FE entire family of tests.
RESULTS
Antibiotic susceptibility profiles
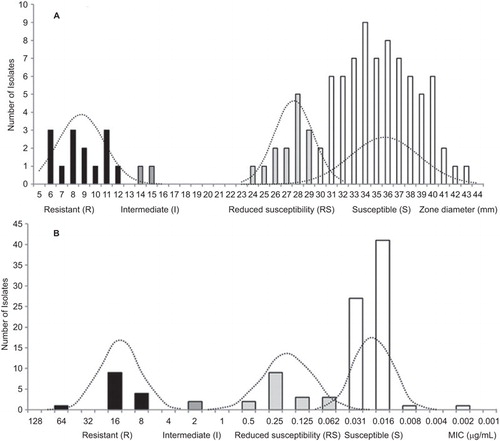
Both the inhibition zones and MIC distributions for E. coli against levofloxacin (, respectively) showed three peaks (three modes) on their histograms. The constructed normal distribution curves showed an intersection between RS and S subpopulations over the range of the inhibition zones from 28 to 32 mm and a MIC of 0.062 µg/mL (, respectively). Cutoff values around the mean of each subpopulation were calculated to be at ±2.0 SDs, which corresponded to 95% of the subpopulation except for the HR suppopulation in which MIC cutoff values were set based on the genotype (). The resultant cutoff values are listed in .
Table 3 Cutoff values of E. coli subpopulations for inhibition zone and MIC distributions
The susceptibility of the 103 E. coli isolates against levofloxacin was distributed as follows: 14 (13.6%) were resistant; two (1.9%) were intermediate; 16 (15.5%) showed reduced susceptibility; and 71 (86.9%) were susceptible.
PCR amplification products and fragment separation
Uniplex PCR reactions resulted in fragments of expected sizes using the primer pairs listed in under the same cycling conditions (). Equimolar concentrations of the primers were then used to perform the multiplex reactions, which resulted in multiple bands; in larger fragments with weak intensities, this was optimized by increasing the primer pair ratio for weak fragments and prolonging the PCR extension time. The products of multiplex reactions were amplified in nearly equal amounts when the conditions listed in were applied (). Interestingly, when fragments close in size (896 (marRA), 952 (gyrA) and 994 (soxSR) bps) moved into the 0.2% agarose region dedicated for extraction, they were more discernible ().
Identification of mutations
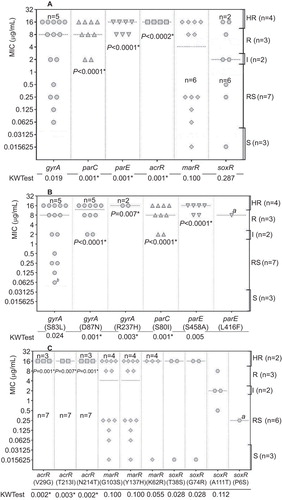
DNA sequencing was performed on a random sample from each subpopulation using the sequencing primers listed in , with a total number of 18 samples sequenced (S (n=1), RS (n=7), I (n=2), R (n=3) and HR (n=5)). The NCBI sequences of the susceptible E. coli K-12 substrains MG1655 and DH10B were included in the analysis. The samples were sequenced only if they were consistently reported to belong to one of the subpopulations by both the disk diffusion and broth microdilution methods. The best qualities and longest chromatogram reads were obtained when uniplex PCR fragments were pooled and copurified (data not shown). The mutation profiles of the sequenced isolates are listed in . All HR isolates showed mutations in all the sequenced genes, whereas all R isolates had mutations in only four genes (three topoisomerases and one efflux regulator), and all I isolates had mutations in only three genes (two topoisomerases and one efflux regulator) ().
Table 4 Topoisomerases and efflux pumps regulators mutations in sequenced E. coli isolates/strains
Interestingly, each of the two HR isolates (ZU14 and ZE29) harbored a total of 13 mutations in six different genes (). It was also observed that R isolates lacked mutations in acrR, and I isolates lacked mutations in acrR, parE and marR. Similarly, RS isolates lacked mutations in acrR, parE, parC, and the double mutation in gyrA, whereas S isolates lacked mutations in acrR, parE, parC and gyrA. None of the sequenced isolates carried mutations in marA (n=16) or soxS (n=14) at any level of resistance ().
On the level of single positions, the following results were observed: (i) none of the R isolates carried mutations in gyrA-R237H, marR-K62R, soxR-T38S, soxR-G74R or any position of acrR; (ii) none of the I isolates carried mutations in gyrA-R237H, soxR-T38S, soxR-G74R or at any position of acrR, parE or marR; (iii) none of the RS isolates harbored mutations in gyrA-R237H, gyrA-D87N, marR-K62R, soxR-T38S, soxR-G74R or any position of acrR, parE or parC; and (iv) none of the S isolates harbored mutations in any position of acrR, parE, parC or gyrA ().
Associations between different mutations and levels of resistance
A total of 20 sets of sequences (18 from the sequencing step in this study and two from NCBI) were subjected to statistical analysis tests. The KW test was first used to pinpoint whether there was an association between any of the levels of resistance with the different mutations. Because the KW test does not identify where the associations occur or how many actually occur, the FE test was then used to locate the associations between specific levels of resistance and specific mutations. Significant associations were found between the following mutations and levels of resistance: (i) the mutations in acrR and HR level, parE and R/HR levels, parC and I/R/HR levels (); (ii) the mutation gyrA-R237H and HR level, gyrA-D87N and I/R/HR levels (); and (iii) mutations in acrR (all positions) and HR level, parE-S458A and R/HR levels, parC S80I and I/R/HR levels (). Corrected P values for individual KW and FE tests are shown in .
DISCUSSION
In this study, we attempted to obtain a panoramic view of the changes in the genotype of E. coli isolates and to pinpoint possible associations between these changes and their corresponding level of resistance to levofloxacin. In previous studies, the mutation pattern of gyrA-S83L/D87N was shown to be related to levofloxacin resistance, whereas the single mutation gyrA-S83L was not.Citation26,Citation27 Faye et al.Citation10 explained that double-mutant enzyme-DNA complexes had a lower affinity for quinolones than wild-type complexes. This explanation is confirmed in this study through the significant association between the double mutation and I/R/HR levels. Moreover, we located a third mutation outside the conventional QRDR, which associated the triple mutant gyrA-S83L/D87N/R237H with HR levels.
Although the role of acrR in establishing a clinically significant HR level could be attributed to the overexpression of the AcrAB efflux pump via acrR mutations, other mechanisms should be considered.Citation7,Citation13,Citation28,Citation29 Similarly, the effect of single and double mutations in parC on resistance were previously reported whereby complementation with the wild-type allele significantly reduced the resistance level.Citation11,Citation13 The contribution of the double mutation soxR-T38S/G74R to resistance may be through the overexpression of SoxS, which could also be overexpressed by other mechanisms.Citation9 No significant association was found between the double mutation marR-G103S/Y137H and any level of resistance. This supports the notion that the contribution of marR mutations to resistance levels is through the accumulation of mutations in other sites in the mar mutants, a phenomenon that was not found in wild-type E. coli isolates.Citation8 The marR-K62R mutation found in some of the HR isolates in this study was previously reported as irrelevant to the loss of MarR function.Citation6
Interestingly, two isolates (found amongst the HR clinical isolates) harbored 13 mutations on six different genes. To the best of our knowledge, this number of mutations has not been previously reported and thus these two isolates are labeled as ‘supermutators’. It should be noted that these isolates were obtained from two different specimens in two different hospitals in Egypt. In fact, the variation in sequence (including silent mutations) and phenotype indicated that all sequenced isolates were from different clones.
There may be additional resistance mechanisms in such supermutator isolates, including the following mechanisms: mutations in mutM, ligB and recG encoding other putative DNA binding enzymes;Citation30 mutations in tolC encoding the outer part of the AcrAB-TolC efflux pump;Citation31 the acquisition of plasmid-borne genes qnr and aac(6′)-Ib-cr that encode drug-blocking and drug-modifying proteins, respectively; the overexpression of quorum-sensing regulator SdiA; mutations in robA encoding the global regulator RobA; and mutations in gyrB encoding the B subunit of the gyrase enzyme.Citation4 Such resistance mechanisms present themselves as interesting locations for investigation in future studies. A previous study showed that QRDR mutations of gyrA, parC and parE, generally had a greater effect on FQ MICs than the acquisition of plasmid-borne genes or overexpression of the efflux pump.Citation32
The novel mutations gyrA-R237H and acrR-V29G associated with the HR isolates require further study to reveal their possible roles in resistance. These mutations may contribute to increased fitness rather than increased levels of resistance.Citation33 The same role could be played by soxR-P6S and parE-L416F, which could not be included in the statistical tests as each was recorded once in the sequenced isolates. Hence, more investigation is needed to sequence a larger number of E. coli isolates with a greater focus on isolates collected from blood samples. This could increase the power of the statistical tests and possibly reveal more significant associations especially for soxR-T38S/G74R and the single mutation gyrA-S83L. The parE-L416F mutation was reported in small ratios of the isolates screened in a previous study.Citation34
The appearance of three modes on the zone diameter and MIC histograms indicated that the collected isolates included non-wild-type isolates that harbored resistance mechanisms. This was previously observed for the MIC distribution of E. coli obtained from The European Committee on Antimicrobial Susceptibility Testing (n=3219) on moxifloxacin showing a pattern with two major peaks and a tendency toward a third peak in between these two peaks. For levofloxacin, the same study set the epidemiological breakpoint for the wild-type population (n=9144) at ≤0.082, whereas European Committee on Antimicrobial Susceptibility Testing reported ≤0.25 and the CLSI reported ≤2 µg/mL.Citation16 Additionally, in this study, the established five-level interpretation criteria included the RS range. Such classifications have been reported before by the CLSI for vancomycin against Staphylococcus aureus.Citation15 In fact, mutations in the gyrA and AcrAB efflux pump regulator genes were found in the RS isolates, which were not distinguished from the S range.
Direct sequencing of products from a multiplex reaction is usually unsuccessful. This could be attributed to the high percentage of the mixture of primers that remains after the purification step. Hence, the separation of the products on an agarose gel would remove the interference of the primer mixture. However, the cut and pooled target fragments would have a high percentage of agarose, which could inhibit subsequent sequencing reactions.Citation21 In this regard, the present study introduced a modification to the method of Ma et al.Citation20 to recover the products from a two-gel system with a minimum agarose percentage and no primer mixture interference. Additionally, we designed multiplex PCR protocols for high-throughput sequencing of the QRDRs of topoisomerase genes gyrA, parC and parE, and/or the multidrug efflux pump AcrAB expression regulation systems acrRAB, marRA, and soxSR (with maximum of six PCR products). Interestingly, a recent study developed a multiplex PCR protocol to detect up to eight plasmid-mediated quinolone-resistance determinants, which enabled the detection of resistance mechanisms in 37% of ciprofloxacin-resistant E. coli isolates.Citation35 Combining this method (to detect plasmid-mediated mechanisms) with the method developed in our study (to detect mechanisms due to chromosomal mutations) could further expand the understanding of quinolone-resistance in future studies.
The authors thank assistant Professor Amr Saeed (Department of Microbiology and Immunology, Faculty of Pharmacy, Suez Canal University, Egypt) for providing facilities and assisting with sequencing, Marwa Essam (Biotechnology Centre, Faculty of Pharmacy, Cairo University, Egypt) for providing the identified enteric E. coli isolates and Miran El-Far (Department of Microbiology and Immunology, Faculty of Pharmacy, Ahram Canadian University, Egypt) for providing the identified uropathogenic E. coli isolates.
- Hooper DC.Mechanisms of action of antimicrobials: focus on fluoroquinolones. Clin Infect Dis2001;32 (Suppl 1): S9–S15.
- Dalhoff A.Global Fluoroquinolone resistance epidemiology and implictions for clinical use. Interdiscip Perspect Infect Dis2012;2012: 976273.
- Khawcharoenporn T, Vasoo S, Ward E, Singh K.High rates of quinolone resistance among urinary tract infections in the ED. Am J Emerg Med2012;30: 68–74.
- Morgan-Linnell SK, Becnel Boyd L, Steffen D et al.Mechanisms accounting for fluoroquinolone resistance in Escherichia coli clinical isolates. Antimicrob Agents Chemother2009;53 235–241.
- Jacoby GA.Mechanisms of resistance to quinolones. Clin Infect Dis2005;41 (Suppl 2): S120–S126.
- Komp Lindgren P, Karlsson A, Hughes D.Mutation rate and evolution of fluoroquinolone resistance in Escherichia coli isolates from patients with urinary tract infections. Antimicrob Agents Chemother2003;47: 3222–3232.
- Wang H, Dzink-Fox JL, Chen M et al.Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: role of acrR mutations. Antimicrob Agents Chemother2001;45: 1515–1521.
- Maneewannakul K, Levy SB.Identification for mar mutants among quinolone-resistant clinical isolates of Escherichia coli. Antimicrob Agents Chemother1996;40: 1695–1698.
- Koutsolioutsou A, Pena-Llopis S, Demple B.Constitutive soxR mutations contribute to multiple-antibiotic resistance in clinical Escherichia coli isolates. Antimicrob Agents Chemother2005;49: 2746–2752.
- Barnard FM, Maxwell A.Interaction between DNA gyrase and quinolones: effects of alanine mutations at GyrA subunit residues Ser83 and Asp87. Antimicrob Agents Chemother2001;45: 1994–2000.
- Heisig P.Genetic evidence for a role of parC mutations in development of high-level fluoroquinolone resistance in Escherichia coli. Antimicrob Agents Chemother1996;40: 879–885.
- Koutsolioutsou A, Martins EA, White DG et al.A soxRS-constitutive mutation contributing to antibiotic resistance in a clinical isolate of Salmonella enterica (Serovar Typhimurium). Antimicrob Agents Chemother2001;45: 38–43.
- Schneiders T, Amyes SGB, Levy SB.Role of AcrR and RamA in fluoroquinolone resistance in clinical Klebsiella pneumoniae isolates from Singapore. Antimicrob Agents Chemother2003;47: 2831–2837.
- Cavaco LM, Frimodt-Møller N, Hasman H et al.Prevalence of quinolone resistance mechanisms and associations to minimum inhibitory concentrations in quinolone-resistant Escherichia coli isolated from humans and swine in Denmark. Microb Drug Resist2008;14: 163–169.
- Clinical and Laboratory Standards Institute.Performance standards for antimicrobial susceptibility testing, 22nd informational supplement. CLSI document M100-S22.Wayne, PA: CLSI, 2012.Available at http://www.indabook.org/preview/8gaxEopPrDb_fkWdELy71yeEBjeFu0sVzLyJn7c_-eM,/M100-S22-Performance-Standards-for-Antimicrobial.html?query=M100-S22-Performance-Standards-for-Antimicrobial
- Kronvall G.Normalized resistance interpretation as a tool for establishing epidemiological MIC susceptibility breakpoints. J Clin Microbiol2010;48: 4445–4452.
- Aly MEA, Essam TM, Amin MA.Antibiotic resistance profile of E. coli strains isolated from clinical specimens and food samples in Egypt. Int J Microbiol Res2012;3: 176–182.
- Aly MEA, Essam TM, Amin MA.Involvement of virulence genes and antibiotic resistance in clinical and food borne diarrheagenic Escherichia coli isolates from Egypt. World J Med Sci2012;7: 276–284.
- Ye J, Coulouris G, Zaretskaya I et al.Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics2012;13: 134.
- Ma H, Difazio S.An efficient method for purification of PCR products for sequencing. BioTechniques2008;44: 921–923.
- Yamaguchi Y, Nimbari S, Ookawara T et al.Inhibitory effects of agarose gel and LB medium on DNA sequencing. BioTechniques2002;33: 282, 284.
- Sanger F, Nicklen S, Coulson AR.DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA1977;74: 5463–5467.
- Sievers F, Wilm A, Dineen D et al.Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol2011;7: 539.
- Arsene S, Leclercq R.Role of a qnr-like gene in the intrinsic resistance of Enterococcus faecalis to floroquinolones. Antimicrob Agents Chemother2007;51: 3254–3258.
- Singh R, Ledesma KR, Chang KT et al.Impact of recA on levofloxacin exposure-related resistance development. Antimicrob Agents Chemother2010;54: 4262–4268.
- Fu Y, Zhang W, Wang H et al.Specific patterns of gyrA mutations determine the resistance difference to ciprofloxacin and levofloxacin in Klebsiella pneumoniae and Escherichia coli. BMC Infect Dis2013;13: 8.
- Li J, Gao X, Luo T et al.Association of gyrA/B mutations and resistance levels to fluoroquinolones in clinical isolates of Mycobacterium tuberculosis. Emerg Microbes Infect2014;3: e19.
- Oethinger M, Kern WV, Jellen-Ritter AS et al.Ineffectiveness of topoisomerase mutations in mediating clinically significant fluoroquinolone resistance in Escherichia coli in the absence of the AcrAB efflux pump. Antimicrob Agents Chemother2000;44: 10–13.
- Webber MA, Talukder A, Piddock LJV.Contribution of mutation at amino acid 45 of AcrR to acrB expression and ciprofloxacin resistance in clinical and veterinary Escherichia coli isolates. Antimicrob Agents Chemother2005;49: 4390–4392.
- Swick MC, Evangelista MA, Bodine TJ et al.Novel conserved genotypes correspond to antibiotic resistance phenotypes of E. coli clinical isolates. PLoS ONE2013;8: e65961.
- Sulavik MC, Houseweart C, Cramer C et al.Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrob Agents Chemother2001;45: 1126–1136.
- Shigemura K, Tanaka K, Yamamichi F et al.Does mutation in gyrA and/or parC or efflux pump expression play the main role in fluoroquinolone resistance in Escherichia coli urinary tract infections?: a statistical analysis study. Int J Antimicrob Agents2012;40: 516–520.
- Marcusson LL, Frimodt-Møller N, Hughes D.Interplay in the selection of fluoroquinolone resistance and bacterial fitness. PLoS Pathog2009;5: e1000541.
- Paltansing S, Kraakman MEM, Ras JMC et al.Characterization of fluoroquinolone and cephalosporin resistance mechanisms in Enterobacteriaceae isolated in a Dutch teaching hospital reveals the presence of an Escherichia coli ST131 clone with a specific mutation in parE. J Antimicrob Chemother2013;68: 40–45.
- Ciesielczuk H, Hornsey M, Choi Vet al.Development and evaluation of a multiplex PCR for eight plasmid-mediated quinolone-resistance determinants. J Med Microbiol2013;62(Pt 12): 1823–1827.