Dear Editor,
The worldwide dissemination of New Delhi metallo-β-lactamase 1 (NDM-1), an Ambler class B metallo-β-lactamase (MBL) conferring resistance to all β-lactams except monobactams, is of great concern for public health. NDM-4, which differs from NDM-1 by a single amino acid substitution (Met154Leu), was demonstrated to possess increased carbapenemase activity.Citation1 As an infrequent blaNDM allele, only several sporadic cases of infections due to NDM-4-producing Escherichia coli have been described in India, Austria, and Europe.Citation1, Citation2, Citation3 At present, only one NDM-4 plasmid (pJEG027; GenBank accession NO KM400601) has been characterized.Citation4 To understand the epidemiology and control the spread of antibiotic-resistant pathogens, we have been investigating carbapenem-resistant Enterobacteriaceae from human patients in China. Previously, we reported a high incidence (33.3%) and endemic spread of NDM-1-positive Enterobacteriaceae in Henan Province.Citation5 Here we describe the first NDM-4-producing E. coli ST410 isolate, which, to our knowledge, is the first case in China, and report the characterization of the blaNDM-4 harboring IncX3 plasmid.
E. coli 14–55 was obtained from a blood culture of a 14-year-old girl hospitalized at The First Affiliated Hospital of Zhengzhou University, Zhengzhou city, Henan province, on 6 June 2014. The girl did not have a history of foreign travel. Susceptibility testing was initially performed by a Vitek 2 system (bioMérieux, Marcy l'Etoile, France), and Minimum Inhibitory Concentrations (MICs) were determined using the broth microdilution method and the agar dilution method (for fosfomycin) following the CLSI guidelines, and the MIC results were interpreted according to the CLSI breakpoints.Citation6 The European Committee on Antimicrobial Susceptibility Testing breakpoints (available at http://www.eucast.org/clinical_breakpoints/) were used for colistin and tigecycline. E. coli 14–55 was resistant to all β-lactams, including imipenem and meropenem, fluoroquinolones, gentamicin and tetracycline, and was susceptible to amikacin, fosfomycin, tigecycline and colistin (Supplementary Table S1). The modified Hodge test and the imipenem-EDTA double-disk synergy test revealed that E. coli 14–55 was an MBL-producer.Citation6 PCR and sequencing were performed to screen for the presence of MBL genes, including blaIMP, blaNDM, and blaVIM; only blaNDM was detected.Citation5 The complete coding sequence (CDS) of blaNDM was amplified with a pair of primers (NDM-F: 5′-GAA TTC GCC CCA TAT TTT TGC; NDM-R: 5′-AAC GCC TCT GTC ACA TCG AAA T) that flanked the blaNDM gene, and DNA sequencing revealed the presence of blaNDM-4 in E. coli 14–55. In addition, different β-lactamase encoding genes, including blaCTX-M-15, blaSHV-12, blaTEM-1 and blaCMY-61, together with the qnrB gene were identified in the NDM-4 positive isolate E. coli 14–55.Citation5
E. coli phylogenetic group typing and multilocus sequence typing (MLST) showed that the isolate E. coli 14–55 belonged to phylogroup A and ST410.Citation5, Citation7 At present, five E. coli STs (India: ST648; Italy and Denmark: ST405; Austria: ST167, ST4450, and ST101) have been identified carrying blaNDM-4 worldwide.Citation1, Citation2, Citation3 Thus, this study represents the first report of ST410 as a carrier of blaNDM-4. Notably, recent publications from Germany and Italy described the clonal expansion of fluoroquinolone (FQ)-resistant CTX-M-15-producing E. coli ST410 in both human and animal populations.Citation8, Citation9 Our previous and current studies also identified E. coli ST410 clinical isolates co-harboring multiple resistance determinants, including blaNDM (blaNDM-1/4), ESBLS (blaCTX-M-15, blaSHV-12/blaTEM-1) and ampC genes (blaCMY-30/blaCMY-61).Citation5 In addition, NDM-5 producing E. coli ST410 was recently detected in Egypt.Citation10 These findings suggest that E. coli ST410 might be an emerging pandemic clone similar to ST131 for the dissemination of CTX-M-15 or NDM encoding genes. This possibility remains to be examined from future surveillance work.
S1-PFGE and PCR-based plasmid replicon typing indicated E. coli 14–55 harbored three different plasmids that belonged to the IncX3, IncFIA and IncFIB incompatibility groups.Citation5, Citation11 (Supplementary Figure S1). Southern hybridization using digoxigenin-labeled blaNDM-1-specific probes (Roche Applied Sciences, Germany) showed that blaNDM-4 was located on an approximately 50 kb plasmid in E. coli 14–55 (Supplementary Figure S1). Although the conjugal transfer of plasmids for E. coli 14–55 failed, the NDM-4 carrying plasmid belonging to the IncX3 incompatibility group was successfully transformed into E. coli DH5α Electro-Cells (TaKaRa, Dalian, China) by electroporation (Supplementary Figure S1). MICs for the transformant (named T55 in this study) are summarized in (Supplementary Table S1). To characterize the IncX3-type NDM plasmid, 12 pairs of primers were designed based on a reference IncX3 plasmid named pNDM-HN380 (NC_019162) for the mapping of the backbone regions (Supplementary Table S2), and the MDR regions were characterized by a primer walking method.
The blaNDM-4 harboring plasmid in E. coli 14–55 (named pEC55-NDM-4) is 54 035 bp in size, with an average G+C content of 49.1%. The sequence data revealed that pEC55-NDM-4 is highly similar (>99.99%) to plasmid pNDM-HN380, which is the first characterized blaNDM-1-harboring IncX3 plasmid of Klebsiella pneumoniae ST483 and was isolated in Hong Kong, with nucleotide changes from G to A at bp 4447, from T to G at 18 179 bp, from G to A at 21 656 bp, and from C to T at 23 844 bp (Figure 1).Citation12 The point mutation at position T18,179 G corresponds to the A460C mutation in blaNDM-1, leading to the occurrence of the blaNDM-4 allele. A comparative plasmid analysis between pEC55-NDM-4 and pJEG027, which is the only completely sequenced NDM-4 plasmid of a K. pneumoniae strain isolated in Austria, provided support for Espedido et al’s hypothesis that pJEG027 might have arisen from a pNDM-HN380-like plasmid ancestor (for example, pEC55-NDM-4) through the events of a different IS5 insertion and an IS26-mediated flanking deletion of cutA1-groL.Citation4 Several recent studies revealed that the blaNDM-4 gene was carried by plasmids belonging to different replicon types, including F, FII, L/M and X3 in E. coli and K. pneumoniae; however, only two partial sequences of IncFII plasmids (accession NO KP826711 and KP826707) and one partial sequence of an IncX3 plasmid (accession NO KP826709) except pJEG027 are available for the genetic context of blaNDM-4.Citation2, Citation3, Citation13 The second completely sequenced blaNDM-4 carrying plasmid pEC55-NDM-4, in our study, is nearly identical to the first characterized blaNDM-1-harboring IncX3 plasmid pNDM-HN380, further suggesting that blaNDM-4 might have emerged on IncX3 plasmids via a point mutation in blaNDM-1. The occurrence of a pNDM-HN380-like plasmid carrying blaNDM-4, in this study, together with recent observations of IncX3-type plasmids carrying different blaNDM alleles, including blaNDM-1 (pNDM-HN380), blaNDM-4 (pJEG027), blaNDM-5 (pNDM_MGR194, pNDM5_0215 and pEc1929), and blaNDM-7 (pEC50-NDM7), in different countries (Austria, India, China and Canada) and different species (K. pneumoniae, E. coli, Serratia marcescens, and Enterobacter hormaechei),Citation4, Citation12, Citation14, Citation15 strongly indicates that IncX3 plasmids, which have a narrow host range in Enterobacteriaceae, might have played a major role in the rapid global dissemination of NDM-type MBLs among Enterobacteriaceae.
Figure 1 Comparison of linear plasmid maps of IncX3 plasmids harboring blaNDM: pNDM-HN380 (blaNDM-1, JX104760), pEC55-NDM-4 (blaNDM-4, KX470734), pJE027 (blaNDM-4, KM400601). Light gray shading indicates homologous regions, while dark gray shading indicates inversely displayed regions of homology. The different arrows indicate the positions, directions of transcription and predicted function of the genes.
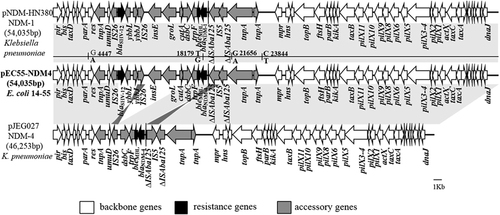
In summary, our study represents the first report of a NDM-4 producing E. coli isolate recovered from a blood culture of a patient without a history of foreign travel. The FQ-resistant E. coli 14–55 with multiple β-lactamase encoding genes, including blaNDM-4, blaCTX-M-15, blaSHV-12, blaTEM-1 and blaCMY-61, belonged to ST410, a potential international clone for the dissemination of CTX-M-15. Further surveillance is thus warranted to monitor the future dissemination of potentially endemic clones of ST410 that harbor blaNDM. The blaNDM-4 carrying IncX3 plasmid characterized in this study is nearly identical to pNDM-HN380 (with blaNDM-1), which was reported in Hong Kong. This finding, together with recent observations of IncX3 plasmids carrying different blaNDM alleles in different countries, further suggests that IncX3 plasmids might have become a common vehicle for the dissemination of different NDM alleles among Enterobacteriaceae worldwide.
Supplementary Information
Download PDF (34.3 KB)Supplementary Information
Download PDF (151.4 KB)Supplementary Information
Download PDF (87 KB)This work was supported by grant 81501782 from the National Natural Science Foundation of China, grant 152300410034 from the Research Project of Basic and Advanced Technology of Henan Province, and grant 15B310014 from the Foundation of Henan Educational Committee. Nucleotide sequence accession number: The sequence of the blaNDM-4 carrying plasmid pEC55-NDM-4 was submitted to the GenBank database under the accession number KX470734.
Supplementary Information for this article can be found on the Emerging Microbes & Infections website (http://www.nature.com/emi)
- NordmannP,BoulangerAE,PoirelL.NDM-4 metallo-beta-lactamase with increased carbapenemase activity from Escherichia coli.Antimicrob Agents Chemother2012; 56:2184–2186.
- CoppoE,Del BonoV,VenturaFet al.Identification of a New Delhi metallo-beta-lactamase-4 (NDM-4)-producing Escherichia coli in Italy.BMC Microbiol2014; 14:148.
- WailanAM,PatersonDL,KennedyKet al.Genomic characteristics of NDM-producing Enterobacteriaceae isolates in Australia and their blaNDM genetic contexts.Antimicrob Agents Chemother2016; 60:136–141.
- EspedidoBA,DimitrijovskiB,van HalSJet al.The use of whole-genome sequencing for molecular epidemiology and antimicrobial surveillance: identifying the role of IncX3 plasmids and the spread of blaNDM-4-like genes in the Enterobacteriaceae.J Clin Pathol2015; 68:835–838.
- QinS,FuY,ZhangQet al.High incidence and endemic spread of NDM-1-positive Enterobacteriaceae in Henan Province, China.Antimicrob Agents Chemother2014; 58:4275–4282.
- Clinical and Laboratory Standards InstitutePerformance Standards for Antimicrobial Susceptibility Testing: Twenty-Fifth Informational Supplement, M07-A10.Wayne, PA, USA: CLSI.2015.
- ClermontO,BonacorsiS,BingenE.Rapid and simple determination of the Escherichia coli phylogenetic group.Appl Environ Microbiol2000; 66:4555–4558.
- FalgenhauerL,ImirzaliogluC,GhoshHet al.Circulation of clonal populations of fluoroquinolone-resistant CTX-M-15-producing Escherichia coli ST410 in humans and animals in Germany.Int J Antimicrob Agents2016; 47:457–465.
- GiufreM,AccogliM,GrazianiCet al.Whole-genome sequences of multidrug-resistant Escherichia coli strains sharing the same sequence type (ST410) and isolated from human and avian sources in Italy.Genome Announc2015; 3:e00757–15.
- GamalD,Fernandez-MartinezM,El-DefrawyIet al.First identification of NDM-5 associated with OXA-181 in Escherichia coli from Egypt.Emerg Microbes Infect2016; 5:e30.
- JohnsonTJ,BielakEM,FortiniDet al.Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae.Plasmid2012; 68:43–50.
- HoPL,LiZ,LoWUet al.Identification and characterization of a novel incompatibility group X3 plasmid carrying blaNDM-1 in Enterobacteriaceae isolates with epidemiological links to multiple geographical areas in China.Emerg Microbes Infect2012; 1:e39.
- KhalifaHO,SolimanAM,AhmedAMet al.NDM-4- and NDM-5-Producing Klebsiella pneumoniae Coinfection in a 6-Month-Old Infant.Antimicrob Agents Chemother2016; 60:4416–4417.
- ChenL,PeiranoG,LynchTet al.Molecular Characterization by Using Next-Generation Sequencing of Plasmids Containing blaNDM-7 in Enterobacteriaceae from Calgary, Canada.Antimicrob Agents Chemother2015; 60:1258–1263.
- ChenD,GongL,WalshTRet al.Infection by and dissemination of NDM-5-producing Escherichia coli in China.J Antimicrob Chemother2016; 71:563–565.
