Abstract
Leptospirosis is a widespread systemic zoonosis, considered as reemerging in certain developing countries. Although the cross agglutinin absorption test is still considered the standard method for Leptospira identification, it presents several disadvantages. The aim of this study was to characterize Leptospira spp. isolated from various hosts by genotyping and broth microdilution susceptibility testing in an attempt to differentiate Leptospira species, serogroups and serovars. Forty-seven isolates were studied. They were previously serotyped, and species confirmation was performed by 16S rRNA sequencing. Single-enzyme amplified fragment length polymorphism (SE-AFLP) and pulsed-field gel electrophoresis (PFGE) analysis enabled the distinction of L. interrogans from L. santarosai, L. meyeri and L. borgpetersenii in two main clusters. Among L. interrogans, it was possible to differentiate into two new clusters the serogroup Icterohaemorrhagiae from the serogroups Canicola and Pomona. L. santarosai isolates presented higher genetic variation than the other species in both techniques. Interestingly, the minimum inhibitory concentration (MIC) cluster analysis also provided Leptospira serogroup differentiation. Further studies are necessary regarding serovar Bananal isolates, as they presented the highest MIC values for most of the antimicrobials tested. All studied techniques successfully distinguished Leptospira species and serogroups. Despite being library-dependent methods, these approaches are less labor intensive and more economically viable, particularly SE-AFLP, and can be implemented in most reference laboratories worldwide to enable faster Leptospira typing.
keywords::
INTRODUCTION
Leptospirosis is a worldwide systemic zoonosis, with higher incidence in tropical climates.Citation1 This disease is caused by bacteria of the Leptospira genus, which are classified into 21 species and nearly 300 serovars organized into 29 serogroups. Among these 21 established species, nine are characterized as pathogenic and frequently isolated from humans and animals; five are considered to be intermediately pathogenic with the ability to infect humans and animals, although less frequently and with variable clinical signs; and seven are considered saprophytic environmental non-pathogenic species.Citation2, Citation3, Citation4, Citation5
Until now, the species identification level was defined by DNA-DNA hybridization data, whereas Leptospira serogroup and serovar classification is based on the expression of the surface antigens.Citation2 Further serological identification is complicated because various serovars can be distributed among different species.Citation6 Although the cross-agglutinin absorption test (CAAT) is still considered the standard method for Leptospira identification, it is highly laborious and expensive because it requires the maintenance of all reference strains and the production of respective antisera.
In this context, molecular methods, with higher discriminatory power and the ability to establish the molecular epidemiology of the isolates and intra-serovar variation, have been applied for Leptospira characterization.Citation3, Citation7, Citation8, Citation9, Citation10 Despite the widespread use of sequence-based molecular methods, with low- to high-throughput scales, they require the use of expensive equipment, rigorously standardized sample preparation protocols and complex bioinformatics analysis. Some genotyping methods of restriction patterns can be quicker and easier to perform; digital analysis enables the standardization and more accurate interpretation of band patternsCitation10 and is more economically viable for the vast majority of researchers from developing countries.
This study aimed to characterize Leptospira spp. isolated from various hosts in Brazil, at different time periods, by single-enzyme amplified fragment length polymorphism (SE-AFLP), pulsed-field gel electrophoresis (PFGE) and broth microdilution for susceptibility profiling, in an attempt to differentiate Leptospira species, serogroups and serovars.
MATERIALS AND METHODS
Bacterial isolates and culture conditions
A total of 47 Leptospira isolates were studied. These isolates originated from the bacterial collection of the Laboratory of Bacterial Zoonosis—University of São Paulo. They were isolated from various hosts, including swine, dog, rat, bovine and human, at different time periods and from different Brazilian states. Forty isolates were previously serotyped at the WHO/FAO/OIE and National Collaborating Centre for Reference and Research on Leptospirosis (Kit Biomedical Research, Amsterdam, the Netherlands) to determine the respective serogroups and serovars.
Cultures were stocked in Fletcher's medium (DIFCO/USA), enriched with 15% rabbit serum and maintained in EMJH medium (DIFCO/USA) at 30 °C until molecular analysis. The L. interrogans serogroup Pomona serovar Pomona reference strain 13A (1937, Australia) and L. interrogans serogroup Icterohaemorrhagiae serovar Copenhageni strain L1.130 (1996, Brazil) were used in this study as internal and quality-control serovars for the experiments.
Molecular typing
Species identification by 16S rRNA sequencing
The species of the isolates that did not belong to L. interrogans were identified by 16S rRNA sequencing. Purified DNA was recovered according to the Boom et al.Citation11 protocol and stored at −20 °C. The 16S rRNA gene amplification was performed as previously described by Morey et al.Citation12 with primer modifications (D1mod—GTT TGA TCC TGG CTC AG; P2mod—GGC TAC CTT GTT ACG ACT T). The amplified fragments were purified using a Illustra GFXTM PCR DNA and Gel Band Purification Kit (GE Healthcare, Little Chalfont, UK) according to the manufacturer’s protocol and sequenced directly at the Human Genome Research Center (University of São Paulo, Brazil). The BIOEDIT Sequence Alignment Editor 7.0.9 Citation13 was used for sequence editing. The DNA sequences from this study were deposited in GenBank under the accession numbers KJ946433—KJ946437, KP739777—KP739784 and KU053945—KU053947.
Single-enzyme amplified fragment length polymorphism (SE-AFLP)
SE-AFLP was performed according to the protocol of McLauchlin et al.Citation14 DNA fragments were detected by electrophoresis at 24 V for 26 h in 2% agarose gel stained with BlueGreen (LGC Biotecnologia, São Paulo, Brazil), and images were captured under UV illumination by a Gel Doc XR System (Bio-Rad Laboratories, Hercules, CA, USA).
Pulsed-field gel electrophoresis (PFGE)
Leptospira seven-day cultures were centrifuged at 5000 rpm for 20 min. The supernatant was discarded, and the pellet was resuspended in 10 mL of PETT IV solution (10 mmol/L Tris-HCl [pH 8.0], and 1 mol/L NaCl, 10 mmol/L EDTA). The bacterial suspension was centrifuged at 1500 rpm for 10 min, the supernatant was discarded, and the pellet was suspended in 1 mL of lysis buffer (1 mol/L NaCl, 10 mmol/L Tris [pH 8.0], 200 mmol/L EDTA, 0.5% sarcosyl, and 0.2% sodium deoxycholate). Agarose SeaKem gold 2% (Cambrex Bio Science Rockland Inc., East Rutherford, NJ, USA) was prepared in 0.5 × Tris Borate EDTA buffer. A volume of 400 μL of the bacterial suspension was heated to 40 °C and added to 20 μL of 100 mg of lysozyme/mL (LGC Biotecnologia, São Paulo, Brazil) and 400 μL of heated 2% agarose solution. The mixture was immediately dispensed into wells and chilled for 10 min at 4 °C. Plugs were placed in 2.5 mL of lysis buffer, and 70 μL of proteinase K (20 mg/mL; LGC Biotecnologia) was added before overnight incubation at 56 °C. The plugs were rinsed once in 1 ml Tris EDTA buffer (10 mmol/L Tris, 1 mmol/L EDTA). The plugs were washed twice with 5 mL of Tris EDTA buffer (10 mmol/L Tris, 1 mmol/L EDTA) for 30 min and then stored in one mL of Tris EDTA buffer (10 mmol/L Tris, 1 mmol/L EDTA) at 4 °C.
DNA was cleaved with NotI enzyme (New England BioLabs, Ipswich, MA, USA), and the PFGE run was performed as described by Galloway and Levett.Citation8 The gels were stained with one(Sybr Safe (Invitrogen Corporation, Carlsbad, CA, USA) for 40 min and photographed under UV transillumination. The DNA fragments were identified using a Lambda DNA-PFGE marker (New England BioLabs, Ipswich, MA, USA), and the images were captured by the Gel Doc XR System (Bio-Rad Laboratories, Hercules, CA, USA).
Broth microdilution susceptibility testing
The susceptibility profiles of the isolates were determined by the antimicrobial minimum inhibitory concentration (MIC) values obtained using the broth microdilution technique. The broth microdilution method was adapted from Murray and Hospenthal’s protocolCitation15 for the use of the Sensititre Standard Susceptibility MIC Plate BOPO6F (TREK Diagnostic Systems/Thermo Fisher Scientific, Waltham, MA, USA). For the inoculum, cultures were grown at 30 °C for seven days and diluted to an optical density, at 420 nm, of 0.32 (approximately 108 CFU/mL), followed by serial dilution using EMJH medium to achieve a final concentration of 2 × 106 CFU/mL (the inoculum was also confirmed by enumeration in a Petroff-Hausser chamber under dark-field microscopy). Fifty microliters of the inoculum were distributed to each well of the Sensititre MIC Plate, and after three days of incubation, 5 μL of 10X alamarBlue (Thermo Fisher Scientific, Waltham, MA, USA) was added to each well. The MIC were assessed visually as the lowest concentration of antibiotics in the wells without color change of alamarBlue on the fifth day of incubation. The susceptibility testing was performed once for each isolate.
Statistical analysis
The phylogenetic analysis of 16S rRNA sequencing was performed using Mega 5.10.Citation16 A dendrogram was constructed using the maximum-likelihood method with the Tamura-3-parameter model. The SE-AFLP and PFGE results were analyzed using the Bionumerics 7.5 software (Applied Maths NV, Saint-Martens-Latem, Belgium). Fingerprint patterns were analyzed by a comprehensive pairwise comparison of restriction fragment sizes, using the Dice coefficient. The mean values obtained from Dice coefficients were employed in UPGMA to generate dendrograms. For SE-AFLP, a cut-off value of 90% of genetic similarity was applied to analyze the resulting clusters; for the PFGE analysis, the isolates were considered to belong to different pulsotypes when differing by four or more bands.Citation17 The discriminatory indexes of both techniques were calculated as described by Hunter and Gaston.Citation18 The MIC cluster analysis was also performed with Bionumerics 7.5 (Applied Maths NV, Saint-Martens-Latem, Belgium) using the Rank correlation method.
RESULTS
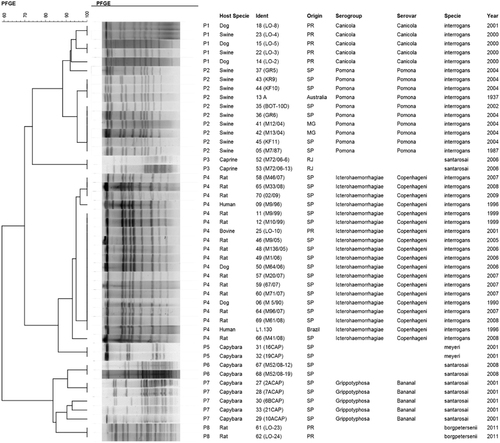
The L. santarosai, L. meyeri and L. borgpetersenii species were confirmed by 16S rRNA sequencing (). SE-AFLP analysis resulted in 15 profiles (A1-A15) comprising the 47 Leptospira isolates (). This technique enabled the distinction of L. interrogans from L. santarosai, L. meyeri and L. borgpetersenii in two main clusters, with over 60% genetic similarity. Among L. interrogans, it was possible to differentiate into two new sub-clusters the serogroup Icterohaemorrhagiae from the serogroups Canicola and Pomona. Serogroup Canicola clustered in profile A5, and the 10 isolates of serogroup Pomona were grouped in four profiles (A1-A4). Although the L. interrogans serogroup Icterohaemorrhagiae clustered together with more than 70% similarity, the 19 isolates were further differentiated into four profiles (A6-A9).
Figure 1 Dendrogram showing the species confirmation of the non-L. interrogans isolates based on 16S rRNA nucleotide sequences. The bootstrap values presented at corresponding branches were evaluated using 500 replicates.
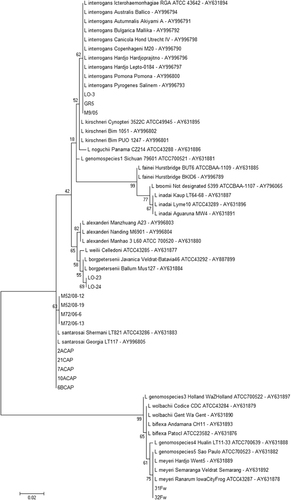
Figure 2 Dendrogram showing the relationships among the SE-AFLP patterns from Leptospira spp. isolates.
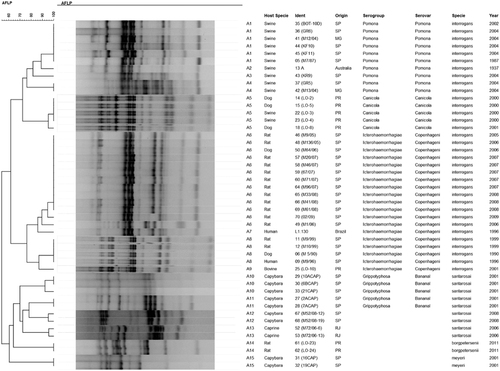
To distinguish them from the L. interrogans cluster, L. santarosai, L. meyeri and L. borgpetersenii isolates could be differentiated from each other at the species level within their cluster. Although the majority of L. santarosai were typed as serogroup Grippotyphosa serovar Bananal and clustered together with over 70% similarity, they presented higher genetic variability than the other isolates and were classified into four profiles (A10-A13). There was no apparent relationship between host species, time period and local of isolation with the SE-AFLP profiles for L. santarosai. The only two L. borgpetersenii isolates were classified as indistinguishable, as were the L. meyeri isolates.
Molecular typing with PFGE also presented the tendency to differentiate L. interrogans from the other Leptospira species studied with more than 60% genetic similarity; however, two L. santarosai isolates (M72/06-6 and M72/06–13) and two L. meyeri isolates (16CAP and 19CAP) presented pulsotypes more closely related to the L. interrogans serogroups Pomona and Icterohaemorrhagiae, respectively (). Despite presenting similar results to SE-AFLP with the distinction of Leptospira species in two main clusters and a tendency to segregate L. interrogans serogroups, PFGE resulted in fewer band patterns with a total of eight pulsotypes (P1-P8) comprising the 47 studied isolates.
Serovar Canicola isolates clustered in P1, whereas P2 corresponded to serovar Pomona isolates and P4 to serogroup Icterohaemorrhagiae. Although PFGE enabled this distinction of L. interrogans serogroups, it also clustered the L. santarosai isolates M72/06-6 and M72/06–13 and the L. meyeri isolates 16CAP and 19CAP (P3 and P5, respectively) in the main group of L. interrogans. Similar to the SE-AFLP results, the L. santarosai isolates presented higher genetic variation in PFGE analysis than the other Leptospira species. The discriminatory indexes obtained for the SE-AFLP and PFGE techniques were 0.89 and 0.78, respectively.
The MIC results for susceptibility testing are shown in . With the exception of penicillin and ampicillin, to which all isolates were susceptible, and trimethoprim/sulfamethoxazole and sulfadimethoxine, which presented higher MIC values, the other antimicrobials tested presented variable MIC values. Cluster analysis based on MIC values also enabled the differentiation of Leptospira serogroups; however, it did not present the same distinction of species as genotyping. Five clusters can be defined in with greater than 50% similarity: the first comprises serogroup Icterohaemorrhagiae isolates with the lowest MIC profile. However, these isolates already present higher values for neomycin, tilmicosin, spectomycin and fluoroquinolones than expected. The increasing MIC values for gentamicin, oxytetracyclin and neomycin determined the second cluster of L. santarosai and L. borgpetersenii and a subcluster of L. interrogans serogroup Canicola isolates with danofloxacin MIC of 4 mg/L.
Figure 4 Dendrogram showing the relationships among the antimicrobial susceptibility profiles of Leptospira spp. isolates. Gray-scale coloring according to increasing MIC values of tested antimicrobials. MIC values presented correspond to single test results.
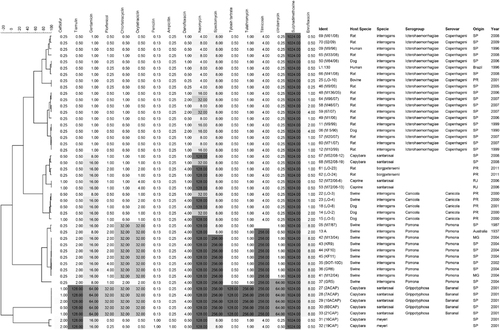
Elevated MIC values for tiamulin, chlortetracycline, oxytetracyclin, tilcomisin and clindamycin, as well as for danofloxacin, neomycin and spectomycin, characterized the L. interrogans serogroup Pomona cluster. The other two clusters comprised the L. santarosai serogroup Grippotyphosa serovar Bananal isolates, which presented the most alarming susceptibility profile, with high MIC values for most of the tested antibiotics with the exception of the β-lactams, and L. meyeri, which also presented a distinct profile with the only isolates with a ceftiofur MIC value of 2 mg/L.
DISCUSSION
Molecular techniques have been applied to Leptospira characterization in an attempt to differentiate species and serovars; however, most Leptospira can be identified only at the species level. Although the practical clinical relevance of serovar identification is still questioned, it is considered an important data source for the study of leptospirosis epidemiology.Citation8 As serovars are associated with specific hosts and even disease severity, their identification can enable the prediction of infection sources and directly assist in controlling the spread of the disease.Citation8, Citation9, Citation19
Our genotyping results, distinguishing L. santarosai, L. borgpetersenii and L. interrogans with greater than 60% genetic similarity, corroborate previous DNA-DNA hybridization data for species definition.Citation19, Citation20 The application of PFGE and SE-AFLP for Leptospira species differentiation represents an alternative for gene sequencing and other molecular methods that are limited to certain pathogenic species, such as the variable number of tandem repeats (VNTRs). SE-AFLP also presents the further advantage of being faster, more economically viable and less troublesome than PFGE and presenting similar results.
Regarding Leptospira serotyping, even though the serovars and serogroups are important epidemiological data, serovars have already been considered a poor indicator of Leptospira genetic relatedness.Citation3 To date, serovar variation has been related to lipopolysaccharide (LPS) structure, specifically the O-antigen, and its biosynthesis locus (rfb cluster).Citation21, Citation22 As molecular typing methods are not directly related to the rfb gene cluster, their application to serovar and serogroup assessment remains controversial.
However, the observed distinction of L. interrogans and L. santarosai serovars by both PFGE and AFLP also corroborates previous reports,Citation6, Citation8, Citation23 such that these methods can be considered valuable tools for Leptospira typing despite their limitations.Citation24 Serogroups must have further genomic differences to allow genotypic distinction. Future studies of comparative genomic analysis may reveal these differences and enhance the molecular serotyping techniques.
Genotyping also enables further epidemiological analysis regarding hosts and infection sources, as we can observe in the SE-AFLP A8 profile that comprises indistinguishable strains of L. interrogans serogroup Icterohaemorrhagiae serovar Copenhageni originating from humans, dogs and rats. This information cannot be assessed by other molecular methods, such as multiple locus sequence typing (MLST), which does not present great correlation with serovars, as it can cluster different serovars in the same sequence type, or isolate origins.Citation8, Citation9
Interestingly, the MIC cluster analysis for the susceptibility testing also provided Leptospira serogroup differentiation. The few previous studies of Leptospira antimicrobial susceptibility focused only on clinical treatment and included a limited number of antimicrobials.Citation25, Citation26, Citation27 Here, we observed differences among the MIC profiles of Leptospira species and of L. interrogans serogroups that are relevant for the clinical and epidemiological assessment of leptospirosis.
From the clinical perspective, even though most isolates were susceptible to penicillin and ampicillin, the classical treatment for leptospirosis, the higher MIC values for tetracyclines, fluoroquinolones, aminoglycosides, tiamulin and spectomycin were not expected, and the veterinary usage of these antibiotics requires attention. The variability of fluoroquinolone susceptibility deserves further attention, as norfloxacin has been indicated as an alternative empirical leptospirosis treatment.Citation28
Regarding the epidemiological assessment, the MIC profile can serve as an alternative tool for Leptospira typing; however, antimicrobial susceptibility is directly related to the host environment and geographical location due to differences in antimicrobial usage worldwide. This relationship could explain the differences between our results and the previous reports by Murray and HospenthalCitation26 and Ressner et al.’s,Citation27 in which no significant variability was observed among the studied strains’ susceptibility profiles. However, further studies are necessary regarding serovar Bananal isolates, which presented the highest MIC values for most antimicrobials, and L. meyeri, which presented higher MIC values for the tested β-lactams.
It should be noted that in this study, only 47 Leptospira isolates were evaluated, comprising field strains exclusively from one country; the small number of L. meyeri and serovar Bananal samples are also drawbacks of the study, despite its noteworthy results. Therefore, larger studies, including isolates from different geographical origins with more representative serogroup/serovar samples, are necessary to allow implementation of the techniques across laboratories worldwide, as well as the assessment of interlaboratory variation.
All three studied techniques have successfully distinguished Leptospira species and serogroups. Although they are classified as library-dependent methods and therefore require a database of reference strains for comparative analysis, they are also less troublesome and more economically viable, particularly SE-AFLP, than the recently enhanced sequencing techniques and can be implemented in most reference laboratories worldwide. Respective limitations and geographical variations must be recognized; nevertheless, SE-AFLP, PFGE and susceptibility testing are alternative methods for Leptospira typing and enhanced epidemiological analysis.
Leptospirosis: budget-friendly pathogen classification
Three techniques offer a cost-effective solution for detecting bacteria responsible for an animal-borne tropical disease. There are 21 species of Leptospira bacteria that can be further sub-classified into hundreds of serovars, but only a fraction cause the disease known as leptospirosis in humans. Identifying that subset generally requires costly genome-sequencing technology, but a team led by Andrea Moreno of the University of São Paulo have identified more economical alternatives. They tested two alternative techniques for DNA analysis, and found that both effectively distinguished Leptospira isolates collected from both humans and animals. A third technique, based on bacterial response to various antibiotics, also proved useful for classification, and revealed one common serovar with unexpectedly low susceptibility to standard drugs. Although less accurate than sequencing, these methods could prove valuable for diagnosis in resource-poor clinical settings.
This study was supported by São Paulo Research Foundation (grants 2011/21502-9; 2011/18290-0 and 2013/17136-2), Coordination for the Improvement of Higher Education Personnel, and National Council for Scientific and Technological Development (CNPq) (Andrea M Moreno and Walter Lilenbaum are CNPq fellows).
- EvangelistaKV,CoburnJ.Leptospira as an emerging pathogen: a review of this biology, pathogenesis and host immune responses.Fut Microbiol2010; 5:1413–1425.
- AdlerB,de la PenaMA.Leptospira and leptospirosis.Vet Microbiol2010; 140:287–296.
- BoonsilpS,ThaipadungpanitJ,AmornchaiPet al.A Single multilocus sequence typing (MLST) scheme for seven pathogenic Leptospira species.PLoS Negl Trop Dis2013; 7:e1954.
- DikkenH,KmetyE.Serological typing methods of Leptospires.In: Bergan T, Norris JR, editors.Methods in Microbiology.New York: Academic Press.1978,pp 268–307.
- VictoriaB,AhmedA,ZuernerRLet al.Conservation of the S10-spc-alpha locus within otherwise highly plastic genomes provides phylogenetic insight into the genus.Leptospira. PLoS One2008; 3:e2752.
- GallowayRL,LevettPN.Evaluation of a modified pulsed-field gel electrophoresis approach for the identification of Leptospira serovars.Am J Trop Med Hyg2008; 78:628–632.
- AhmedN,ManjulataDS,ValverdeMet al.Multilocus sequence typing method for identification and genetic classification of pathogenic Leptospira species.Ann Clin Microbiol Antimicrob2006; 5:1–10.
- GallowayRL,LevettPN.Application and validation of PFGE for serovar identification of Leptospira clinical isolates.PLoS Negl Trop Dis2010; 4:1–7.
- RomeroEC,BlancoRM,GallowayRL.Analysis of Multilocus Sequence Typing for Identification of Leptospira Isolates in Brazil.J Clin Microbiol2011; 49:3940–3942.
- ThaipadungpanitJ,WuthiekanunV,ChierakulWet al.A dominant clone of Leptospira interrogans associated with an outbreak of human leptospirosis in Thailand.PLoS Negl Trop Dis2007; 1:e56.
- BoomR,SolCJA.Salimans MMM. Rapid and simple method for purification of nucleic acids.J Clin Microbiol1990; 28:495–503.
- MoreyRE,GalloweyRL,BraggSLet al.Species-specific identification of Leptospiraceae by 16S rRNA gene sequencing.J Clin Microbiol2006; 44:3510–3516.
- HallTA.BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT.Nucleic Acids Sym Series1999; 41:95–98.
- McLauchlinJ,RipabelliG,BrettMM,ThrelfallEJ.Amplified fragment length polymorphism (AFLP) analysis of Clostridium perfringens for epidemiological typing.Int J Food Microbiol2000; 56:21–28.
- MurrayCK,HospenthalDR.Broth microdilution susceptibility testing for Leptospira spp.Antimicrob Agents Chemother2004; 48:1548–1552.
- TamuraK,PetersonD,PetersonNet al.MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony method.Mol Biol Evol2011; 28:2731–2739.
- Van BelkumA,TassiosPT,DijkshoornLet al.Guidelines for the validation and application of typing methods for use in bacterial epidemiology.Clin Microbiol Infect2007; 13:1–46.
- HunterPR,GastonMA.Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity.J Clin Microbiol1988; 26:2465–2466.
- LevettPN.Leptospirosis.Clin Microbiol Rev2001; 14:296–326.
- YasudaPH,SteigerwaltAG,SulzerKRet al.Deoxyribonucleic acid relatedness between serogroups and serovars in the family Leptospiraceae with proposals for seven new Leptospira species.Int J Syst Bacteriol1987; 37:407–415.
- FaineS,AdlerB,BolinC,PerolatP.Leptospira and Leptospirosis.Melbourne: MediSci.1999.
- KalambahetiT,BulachDM,RajakumarK,AdlerB.Genetic organization of the lipopolysaccharide O-antigen biosynthetic locus of Leptospira borgpetersenii serovar hardjobovis.Microb Pathog1999; 27:105–117.
- VijayachariP,AhmedN,SugunanAPet al.Use of fluorescent amplified fragment length polymorphism for molecular epidemiology of leptospirosis in India.J Clin Microbiol2004; 42:3575–3580.
- SlackAT,DohntMF,SymondsML,SmytheLD.Development of a Multiple-Locus Variable Number of Tandem Repeat Analysis (MLVA) for Leptospira interrogans and its application to Leptospira interrogans serovar Australis isolates from Far North Queensland, Australia.Ann Clin Microbiol Antimicrob2005; 30:4–10.
- HospenthalD,MurrayCK.In vitro susceptibilities of seven Leptospira species to traditional and newer antibiotics.Antimicrob Agents Chemother2003; 47:2646–2648.
- MurrayCK,HospenthalDR.Determination of susceptibilities of 26 Leptospira sp. serovars to 24 antimicrobial agents by a broth microdilution technique.Antimicrob Agents Chemother2004; 48:4002–4005.
- RessnerRA,GriffithME,BeckiusMLet al.Antimicrobial susceptibilities of geographically diverse clinical human isolates of Leptospira.Antimicrob Agents Chemother2008; 52:2750–2754.
- ChakrabortyA,MiyaharaS,VillanuevaSYAM,GlorianiNG,YoshidaS.In vitro sensitivity and resistance of 46 Leptospira strains isolated from rats in the Philippines to 14 antimicrobial agents.Antimicrob Agents Chemother2010; 54:5403–5405.