Abstract
Emerging Microbes and Infections (2016) 5, e49; doi:10.1038/emi.2016.49; published online 25 May 2016
Dear Editor,
The epidemiology of Candida species-associated invasive fungal infections is evolving, and many uncommon Candida species have recently emerged as etiologic agents of bloodstream and other invasive infections. This emergence is attributable to the use of antifungal drugs, such as azoles, for prophylaxis and echinocandins among high-risk populations. Notably, these species exhibit decreased in vitro susceptibility to the antifungals used for therapy.Citation1 In the last few years, clinical treatment failures for Candida haemulonii infections associated with resistance to amphotericin B (AMB) and reduced susceptibility to azoles and echinocandins have been reported.Citation2, Citation3, Citation4 Members of the Candida haemulonii species complex are uncommon yeasts that cause bloodstream and deep-seated infections, and consist of two genotypically distinguishable species, that is, C. haemulonii and C. duobushaemulonii, and a variety, C. haemulonii var. vulnera.Citation2 These species and other relatives of C. haemulonii, that is, Candida auris and Candida pseudohaemulonii, cannot be differentiated by the commercial yeast identification methods used in microbiology laboratories; thus, the true distributions of C. haemulonii and sibling species remain unknown.Citation5
Although the prevalence of uncommon Candida species vary geographically, infections due to C. haemulonii are primarily reported in South America, Asia, the Middle East and Europe.Citation2, Citation3, Citation4, Citation5, Citation6, Citation7, Citation8 Despite their emergence as opportunistic yeasts in several countries, there is a paucity of data regarding the genetically related C. haemulonii species complex from clinical sources. In addition, unlike its sibling species, C. auris, which is known for clonal endemicity in hospitals, no information regarding the genetic diversity of the C. haemulonii species complex is available.Citation5 Herein, we describe the emergence of the C. haemulonii species complex in three hospitals in India. Furthermore, multilocus phylogenetic analyses were performed to investigate the species/varieties in the clinical C. haemulonii species complex, and amplified fragment-length polymorphism (AFLP) fingerprinting was applied to determine their genetic diversity. AFLP was first described 20 years ago and is known to have high discriminatory power due to the generation of many informative genetic markers.Citation9 In addition, the antifungal susceptibility profile of the C. haemulonii species complex was determined.
Clinical isolates were collected from 2010 to 2015 from individual patients in two tertiary care hospitals in Delhi and a solitary hospital in Kochi, Kerala, in South India. The isolates were presumptively identified as C. haemulonii with the VITEK2 system (Biomerieux, Marcy l'Etoile, France) in local hospitals. However, matrix-assisted laser desorption ionization–time-of-flight mass spectrometry (MALDI–TOF MS; Bruker-Daltonics, Bremen, Germany) accurately identified the species and variety in the C. haemulonii species complex. These species included C. duobushaemulonii (n=8), C. haemulonii (n=6) and C. haemulonii var. vulnera (n=1). The species’ morphologies were studied on Sabouraud dextrose agar, CHROMagar Candida medium (Becton-Dickinson, Baltimore, MI, USA) and rice Tween-80 agar, and their growth characteristics were examined at 37 °C and 42 °C. Antifungal susceptibility testing with fluconazole (FLU), itraconazole (ITC), voriconazole (VRC), isavuconazole (ISAV), posaconazole (POS), AMB, 5-flucytosine (FC), caspofungin (CAS), micafungin (MFG) and anidulafungin (AFG) was performed using both the M27-A3 Clinical and Laboratory Standards Institute (CLSI) broth microdilution methodCitation10 and the AST-YS07 card of the VITEK2. Furthermore, to determine the multilocus phylogeny, four loci, including the internal transcribed spacer region (ITS), D1/D2, RPB1 and RPB2, were sequenced.Citation2 A neighbor-joining (NJ) phylogenetic tree based on the concatenated sequences of the four loci was constructed using MEGA version 6 (http://www.megasoftware.net/). In addition, as previously described, an AFLP dendrogram was generated with Bionumerics v6.6 (Applied-Maths, Sint-Martens-Latem, Belgium) using the standard Pearson and unweighted pair group method with averages (UPGMA).Citation11
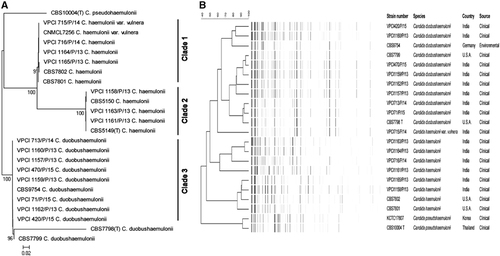
GenBank BLAST searches of the ITS region and MALDI–TOF MS confirmed the presence of 15 isolates belonging to C. duobushaemulonii (n=8), C. haemulonii (n=6) and C. haemulonii var. vulnera (n=1). The ITS sequences of all of the isolates exhibited 100% homologies with the type and reference strains of the respective species in GenBank. The isolates originated from the deep-seated tissue or bone obtained during surgical debridement (n=9), blood (n=5) and BAL (n=1). The isolates exhibited budding yeast cells with pseudohyphae and developed a dark pink color in CHROMagar Candida medium after 48 h of incubation at 37 °C. The isolates grew well at 37 °C, whereas no growth was observed at 42 °C. The NJ tree consisted of three major clades (Figure 1). Clades 1 and 2 contained C. haemulonii (n=3 each) and a solitary C. haemulonii var. vulnera isolate in addition to the type and reference strains, and suggested strain variation. The similarity percentages among the C. haemulonii isolates were 79%–89%. However, all of the C. duobushaemulonii clustered together in clade 3. Notably, and as previously reported, the currently available loci (ITS, LSU, RPB1 and RPB2) possessed low discriminatory power for describing the intra-species genetic diversity of C. haemulonii, C. haemulonii var. vulnera and C. duobushaemulonii.Citation2 Thus, the inclusion of better polymorphic genes for multilocus sequence analysis is warranted for investigations of the clonal transmissions of these species. The AFLP fingerprint analysis revealed markedly variable banding patterns with ~50 bands/strain comprising 25–425 bp overall.
High geometric mean (GM) minimum inhibitory concentrations (MICs) for FLU (CLSI 12.7 mg/L; VITEK2 24.2 mg/L) and AMB (CLSI 14.6 mg/L; VITEK2 6.6 mg/L) were observed via both the CLSI and VITEK2 methods. In contrast, all isolates exhibited low CLSI GM MICs for ISA (0.02 mg/L), POS (0.08 mg/L), ITC (0.3 mg/L) and VRC (0.134 mg/L) with the exception of a solitary C. haemulonii isolate that exhibited a high MIC for VRC (4 mg/L). Notably, a significant twofold greater CLSI MIC for FLU was observed for C. haemulonii (GM 26.2 mg/L) than for C. duobushaemulonii (GM 6.72 mg/L; Supplementary Table S1). In addition, all of the isolates exhibited low GM MICs for CAS (CLSI 0.15 mg/L; VITEK2 0.30 mg/L), MFG (CLSI 0.33 mg/L; VITEK2 0.10 mg/L) and AFG (CLSI 0.5 mg/L), with the exception being solitary isolates of C. haemulonii var. vulnera and C. duobushaemulonii, which exhibited high MICs (1 mg/L) against all of the echinocandins and high MICs (1 mg/L) for AFG and MFG, respectively, by CLSI. Furthermore, significantly lower (Mann-Whitney P<0.05) GM MICs for VRC, CAS and FC, and higher GM MICs for AMB and MFG were observed by CLSI compared with VITEK2. However, the two methods exhibited good agreement (80%–100%) for AMB, FLU, CAS and MFG, but a low agreement (20%) was noted for VRC. Therefore, standardization of VITEK2 via testing with a larger number of C. haemulonii isolates is warranted to validate the use of the VITEK2 system as an alternative method for routine use in laboratories for susceptibility testing.
The clinical features and outcomes of the 15 patients were retrieved from records (Supplementary Table S2). Nine patients had chronic foot infections, and surgically debrided bone and soft tissues yielded C. haemulonii isolates that included C. haemulonii var. vulnera in five patients and C. duobushaemulonii in four. All nine patients had the major risk factor of uncontrolled diabetes and underwent foot amputations. Two of the nine patients also received azole antifungals. Among the five candidemia patients, two had acute myeloid leukemia and the remaining three patients had the underlying conditions of renal transplant and abdominal surgery. Breakthrough candidemia while on azole prophylaxis (FLU=3 and VRC=1) was noted in four patients. VRC therapy was initiated in three candidemia patients and two were successfully managed. The 28-day all-cause mortality among the candidemia patients was 60%. Overall, the patients had at least three predisposing risk factors for candidiasis as previously reported,Citation12 and these risk factors included the following: 15 (100%) were on broad-spectrum antibiotics, ten (66%) had diabetes mellitus, six (40%) had chronic kidney conditions, five (33.3%) had coronary artery disease, four (27%) had peripheral occlusive vascular disease, three (20%) had malignancies, two (13%) had undergone renal transplant and one had undergone abdominal surgery.
Figure 1 (A) Neighbor-joining phylogenetic tree based on the ITS, D1/D2, RPB1 and RPB2 sequences with 2000 bootstrap replications using MEGA version 6. (B) AFLP dendrogram using UPGMA in combination with the Pearson correlation coefficients of the Candida haemulonii species complex and the reference strains (C. duobushaemulonii CBS9754, CBS7799 and CBS7798T; C. haemulonii CBS7801 and CBS7802; and C. pseudohaemulonii CBS 10004T and KCTC17807). The scale bar indicates the percentage similarity. amplified fragment-length polymorphism, AFLP; internal transcribed spacer region, ITS; unweighted pair group method with averages, UPGMA.
Worldwide, the uncommon Candida species that cause infections are not well characterized.Citation1, Citation13 The present study reports the large series of candidiasis cases due to the C. haemulonii species complex with clinical and microbiological data that provide novel insights into this yeast. Unlike its sibling species, C. auris, which is widely prevalent in Indian hospitals, C. haemulonii accounts for only 0.8% of Candida species. Notably, previous reports regarding the occurrence of C. haemulonii in India are misleading because the identifications were performed with conventional or commercial identification systems.Citation14 Recently, Kathuria et al.Citation15 reported that of 102 C. haemulonii isolates identified with the VITEK2 system in five centers in India, 88.2% (n=90) were subsequently confirmed as C. auris by ITS sequencing.
Notably, a high frequency of infections due to the C. haemulonii species complex was observed in the patients with diabetes mellitus (66.6%). Furthermore, as previously reported, the C. haemulonii isolates exhibited elevated MICs to AMB and FLU.Citation2, Citation3 In addition, the high MICs to echinocandins noted in two isolates (C. haemulonii var. vulnera and C. duobushaemulonii) are worrisome. Thus far, two previous studies have reported high MICs for echinocandins.Citation2, Citation4 In conclusion, this study highlights the significance of C. haemulonii species in chronic foot infections and candidemia, and the cases of the latter were associated with breakthrough infections, specifically in the patients on FLU therapy. Thus, correct identifications and determinations of the antifungal susceptibilities of C. haemulonii isolates are warranted to guide antifungal therapy. Because the clinical experience with C. haemulonii infections is limited, optimal treatments have not yet been defined.Citation16 However, the in vitro activities of VRC, POS and ISAV suggest that they can be used as effective treatment options.
GenBank nucleotide sequence accession numbers
ITS, KP862806-KP862817 and KU361136-KU361138; D1/D2, KU361120-KU361134; RPB1, KU361809-KU361103; and RBP2, KU3361104-KU361118.
Supplementary Table s1
Download PDF (421.7 KB)Supplementary Table s2
Download PDF (255.4 KB)- JungDS,FarmakiotisD,JiangYet al.Uncommon Candida species fungemia among cancer patients, Houston, Texas, USA.Emerg Infect Dis2015; 21:1942–1950.
- Cendejas-BuenoE,KoleckaA,Alastruey-IzquierdoAet al.Reclassification of the Candida haemulonii complex as Candida haemuloniiC. haemulonii group I), C. duobushaemulonii sp. nov.C. haemulonii group II), and C. haemulonii var. vulnera var. nov.: three multiresistant human pathogenic yeasts.J Clin Microbiol2012; 50:3641–3651.
- RuanSY,KuoYW,HuangCTet al.Infections due to Candida haemulonii: species identification, antifungal susceptibility and outcomes.Int J Antimicrob Agents2010; 35:85–88.
- RamosLS,Figueiredo-CarvalhoMH,BarbedoLSet al.Candida haemulonii complex: species identification and antifungal susceptibility profiles of clinical isolates from Brazil.J Antimicrob Chemother2015; 70:111–115.
- ChowdharyA,SharmaC,DuggalSet al.New clonal strain of Candida auris, Delhi, India.Emerg Infect Dis2013; 19:1670–1673.
- MuroMD,MottaFdeA,BurgerMet al.Echinocandin resistance in two Candida haemulonii isolates from pediatric patients.J Clin Microbiol2012; 50:3783–3785.
- KimMN,ShinJH,SungHet al.Candida haemulonii and closely related species at 5 university hospitals in Korea: identification, antifungal susceptibility, and clinical features.Clin Infect Dis2009; 48:e57–e61.
- KhanZU,Al-SweihNA,AhmadSet al.Outbreak of fungemia among neonates caused by Candida haemulonii resistant to amphotericin B, itraconazole, and fluconazole.J Clin Microbiol2007; 45:2025–2027.
- VosP,HogersR,BleekerMet al.AFLP: a new technique for DNA fingerprinting.Nucleic Acids Res1995; 23:4407–4414.
- Clinical Laboratory Standards Institute (CLSI)Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts (approved standard M27-A3).3rd edn.Wayne, PA, USA: National Committee for Clinical Laboratory Standards; 2008.Available athttp://shop.clsi.org/microbiology-documents/M27.html.
- ChowdharyA,Anil KumarV,SharmaCet al.Multidrug-resistant endemic clonal strain of Candida auris in India.Eur J Clin Microbiol Infect Dis2014; 33:919–926.
- KullbergBJ,ArendrupMC.Invasive candidiasis.N Engl J Med2015; 373:1445–1456.
- PfallerMA,DiekemaDJ,GibbsDLet al.Results from the ARTEMIS DISK Global Antifungal Surveillance Study, 1997 to 2007: a 10.5-year analysis of susceptibilities of Candida species to fluconazole and voriconazole as determined by CLSI standardized disk diffusion.J Clin Microbiol2010; 48:1366–1377.
- OberoiJK,WattalC,GoelNet al.Non-albicans Candida species in blood stream infections in a tertiary care hospital at New Delhi, India.Indian J Med Res2012; 136:997–1003.
- KathuriaS,SinghPK,SharmaCet al.Multidrug resistant Candida auris misidentified as Candida haemulonii: Characterization by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) and DNA sequencing and its antifungal susceptibility profile variability by Vitek 2, CLSI Broth Microdilution, and Etest method.J Clin Microbiol2015; 53:1823–1830.
- ArendrupMC,BoekhoutT,AkovaMet al.ESCMID and ECMM joint clinical guidelines for the diagnosis and management of rare invasive yeast infections.Clin Microbiol Infect2014; 20:76–98.
