Abstract
Adhesion is an important virulence trait of Vibrio alginolyticus. Bacterial adhesion is influenced by environmental conditions; however, the molecular mechanism underlying this effect remains unknown. The expression levels of flrA, flrB and flrC were significantly downregulated in adhesion-deficient V. alginolyticus strains cultured under Cu2+, Pb2+, Hg2+ and low-pH stresses. Silencing these genes led to deficiencies in adhesion, motility, flagellar assembly, biofilm formation and exopolysaccharide (EPS) production. The expression levels of fliA, flgH, fliS, fliD, cheR, cheV and V12G01_22158 (Gene ID) were significantly downregulated in all of the RNAi groups, whereas the expression levels of toxT, ctxB, acfA, hlyA and tlh were upregulated in flrA- and flrC-silenced groups. These genes play a key role in the virulence mechanisms of most pathogenic Vibrio species. Furthermore, the expression of flrA, flrB and flrC was significantly influenced by temperature, salinity, starvation and pH. These results indicate that (1) flrA, flrB and flrC are important for V. alginolyticus adhesion; (2) flrA, flrB and flrC significantly influence bacterial adhesion, motility, biofilm formation and EPS production by controlling expression of key genes involved in those phenotypes; and (3) flrA, flrB and flrC regulate adhesion in the natural environment with different temperatures, pH levels, salinities and starvation time.
Emerging Microbes & Infections (2016) 5, e85; doi:10.1038/emi.2016.82; published online 3 August 2016
INTRODUCTION
Vibrio alginolyticus is an important opportunistic pathogen and causes vibriosis, one of the most prevalent diseases in maricultured animals and seafood consumers.Citation1 The large yellow croaker (Pseudosciaena crocea) is one of the most economically important maricultured fish species in southeast China.Citation2 V. alginolyticus readily causes vibriosis in cultured P. crocea, which results in severe economic losses in the warm-water season.Citation3
Bacterial adhesion to host surfaces is one of the initial steps in the infection process.Citation4, Citation5 Host mucus is abundantly found on the surface of the skin, gills and gut lining; therefore, it is the first site of interaction between the pathogen and its host.Citation6 Thus, research into adhesion of pathogenic bacteria to host mucus has gained much interestCitation7, Citation8 and the genes involved in bacterial adhesion are starting to be explored.Citation9, Citation10 However, to our knowledge, the studies on the genetic regulation of adhesion in pathogenic bacteria conducted thus far are limited, and the genetic regulatory network is still largely unknown.
Bacterial adhesion of pathogenic V. alginolyticus to the mucus of P. crocea has been studied by our laboratory for several years. The characteristics of V. alginolyticus adhesion were found to correlate with its epidemiologic characteristics,Citation11, Citation12 and bacterial adhesion was found to be influenced by environmental factors such as temperature, pH and salinity, among others.Citation13 To explore the mechanisms by which environmental factors influence bacterial adhesion, pathogenic V. alginolyticus was subcultured under nine different stress conditions. Of these, Cu2+, Pb2+, Hg2+ and low pH were found to reduce the adherence of V. alginolyticus, whereas high pH, low or high salinity, and low or high temperature did not significantly affect adhesion.Citation14 Next, the adhesion-attenuated culture and a control culture were subjected to RNA-seq.Citation14 The RNA-seq results indicated a role for the flagellar assembly genes and three noncoding RNAs, and these were further studied to determine their role in bacterial adhesion.Citation3, Citation15
The flagellum has been shown to be involved in bacterial adhesion and invasion of host cells, as it is used in motility, chemotaxis and as an adhesin.Citation16 The synthesis and assembly of the flagellum is a tightly regulated process.Citation17 flrA, rpoN, flrB and flrC are part of a two-component systemCitation18 that is considered a key prerequisite for bacterial pathogenesis.Citation19 The histone-like nucleoid-structuring protein and the general stress-response regulator RpoS could enhance the transcription of flrA and rpoN.Citation20 Both of FlrA and RpoN-bound RNA polymerase could activate the transcription of class II flagellar genes.Citation21 The flrA transcription start is 68 bp upstream from the flrA translation start site, and DNA sequence upstream of the transcription start contains a σ70 promoter sequence (TTGACA-N14-TGGCACTTT).Citation22 FlrA could directly bind to the flrBC promoter. The binding site is 73–53 bp from the transcription start site (consisted of an inverted repeat sequence of ATTG(A/G)C), which is 24 bp upstream of the translation start site of the flrBC operon.Citation23 FlrB and FlrC could activate the transcription of class III genes.Citation21 flrA, flrB and flrC were first identified in V. cholerae and shown to be required for flagellar synthesis.Citation21 Since then, only a few related studies were performed and mainly focused on the role of flrA, flrB and flrC in flagellar synthesis.Citation17, Citation24 However, the role they and their downstream genes have in the virulence mechanisms of most pathogens is poorly understood and remains to be elucidated.
The results of RNA-seq by our laboratory showed that flrA, flrB and flrC were significantly downregulated under several stress conditions (Cu2+, Pb2+, Hg2+ and low pH). As V. alginolyticus adhesion was also significantly inhibited by these four stress conditions, these genes were hypothesized to have a role in bacterial adhesion. Here, we examine the genes that contribute to adhesion of pathogenic V. alginolyticus. The purpose of our study was to investigate (i) the relationship between flrA, flrB, flrC and V. alginolyticus adhesion; (ii) how these genes affect adhesion; and (iii) whether these genes participate in the regulation of adhesion in natural conditions.
MATERIALS AND METHODS
Bacterial strain and culture conditions
Pathogenic V. alginolyticus (ND-01) was isolated from a spontaneously infected P. crocea and previously identified as pathogenic by subsequent artificial infection.Citation25 Bacteria were stored in physiological saline with 10% glycerol at −80 °C. Bacteria were grown in LB20 medium (Luria-Bertani medium plus 2% NaCl)Citation26 with shaking at 220 rpm.
RNA sequencing (RNA-Seq) results were confirmed by quantitative reverse transcriptase real-time polymerase chain reaction (qRT-PCR). V. alginolyticus was stressed with 50 mg/L Cu2+, 100 mg/L Pb2+, 50 mg/L Hg2+ or low pH (HCl was used to lower the pH to 5). As a control, V. alginolyticus was also incubated under normal conditions (LB20 broth with 2% NaCl, pH=7). Each treatment consisted of six independent biological replicates (three technical replicates within each).
To investigate the effects of different temperatures, V. alginolyticus was cultured in LB20 broth (supplemented with 2% NaCl, pH=7) at 4°C, 15°C, 28°C, 37°C and 44 °C. To investigate the effects of different pH levels, V. alginolyticus was cultured in LB20 broth (supplemented with 2% NaCl) adjusted to a pH of 5, 6, 7, 8 or 9.Citation11 To investigate the effects of different salinities, V. alginolyticus was cultured in LB20 broth (pH=7) with 0.8%, 1.5%, 2.5%, 3.5% or 4.5% NaCl.Citation11 To investigate the effects of starvation, V. alginolyticus was cultured in normal phosphate-buffered saline (PBS) at 28 °C for one, three, five and seven days.Citation27 Each treatment consisted of six independent biological replicates (three technical replicates within each). After harvesting and re-suspending, RNA was extracted from the bacteria, and reverse transcription and qRT-PCR were performed.
Escherichia coli SM10 was purchased from TransGen Biotech (Beijing, China). Cultures were grown in LB20 broth (220 rpm, 37 °C) or on LB20 plates (with 1.8% agar, 37 °C).
Functional classification and enrichment analysis of differentially expressed genes
To annotate identified differentially expressed genes (DEGs), the Blast2GO program was used to obtain Gene Ontology (GO) annotations for each unigene. After obtaining the GO annotation of each unigene, the WEGO software (BGI, Shenzhen, China) was used to determine GO functional categories of all unigenes and to illustrate the species layout of unigene functions in a macroscopic view. The Bonferroni correction was used to calculate the P-value. Adjusted P⩽0.05 was used as a threshold. GO terms satisfying this criterion were defined as notably enriched GO terms from the DEGs.
Kyoto encyclopedia of genes and genomes (KEGG) pathway annotations were obtained using the Blastall software (NCBI, Bethesda, MD, USA; http://www.ncbi.nlm.nih.gov/BLAST/) and the KEGG (http://www.genome.jp/kegg/) database. Q-values were defined as the false discovery rate analog of the P-value. Pathways with Q⩽0.05 were deemed significantly enriched in DEGs.
Transient gene silencing
Negative control short interfering RNA (siRNA) and treatment siRNA sequences (targeting at 136 bp–155 bp downstream from the flrA translation start site, 728 bp–747 bp downstream from the flrB translation start site and 935 bp–954 bp downstream from the flrC translation start site) were chemically synthesized by GenePharma Co. Ltd (Shanghai, China) and are listed in Supplementary Table S1.
Electrotransformation of bacteria was performed following the method of Lancashire et al.Citation28 Overnight cultures of V. alginolyticus were inoculated into fresh LB20 broth (1:100 v/v) and incubated until the OD600 reached 0.3–0.5. Bacterial cells were harvested by centrifugation at 4000 g for 10 min at 4 °C, thoroughly washed twice with ice-cold sterile water at 4 °C and subsequently washed once in ice-cold 10% washing buffer (10% redistilled glycerol, 90% sterile water, v/v). Finally, the competent cells were re-suspended in 1 mL ice-cold 10% glycerol and stored at −80 °C for a short time.
V. alginolyticus-competent cells were electroporated using a Bio-Rad MicroPulser (Bio-Rad Laboratories Inc., Hercules, CA, USA) following the method of Huang et al.Citation15 Two microliters of 20 μM siRNA was vigorously mixed into 100 μL of electrocompetent cells. After incubating on ice for 30 min, the mixture of cells/siRNA was transferred into an electroporation cuvette and electroporated at 1.8 kV for 6 ms. Then, 900 μL of room temperature LB20 medium was immediately added, and the cells were shaken for 1 h, 6 h, 12 h and 24 h at 28 °C for RNA extraction and qRT-PCR.
Stable gene silencing
The stable gene-silencing assay for V. alginolyticus was implemented based on Darsigny et al.Citation29 and Choi and Schweizer.Citation30 Overnight cultures of V. alginolyticus were collected by centrifugation and treated as described in the transient gene-silencing section. Three short hairpin RNA sequences targeting at 136 bp–155 bp downstream from the flrA translation start site, 728 bp–747 bp downstream from the flrB translation start site and 935 bp–954 bp downstream from the flrC translation start site were designed by Shanghai Generay Biotech Co., Ltd (Shanghai, China; Supplementary Table S2). The annealed oligonucleotides were ligated with pACYC184 vectors, which were harvested from BamHI and SphI double digestion via T4 DNA ligase (TaKaRa, Kusatsu, Japan) according to the manufacturer’s recommendations. Competent E. coli SM10 cells were transformed with the recombinant pACYC184 vectors via heat-shock and then transferred by conjugation from E. coli SM10 to V. alginolyticus. The empty plasmid pACYC184 was also transferred into cells and used as the control strain. The stable silenced clones were screened using LB20 medium containing chloramphenicol (34 μg/mL).
Mucus preparation
P. crocea care and use were conducted in strict accordance with the recommendations in the ‘Guide for the Care and Use of Laboratory Animals’ set by the National Institutes of Health. The protocol was approved by the Animal Ethics Committee of Xiamen University (Acceptance NO XMULAC20120030).
Healthy P. crocea were obtained from marine cultured cages in the city of Ningde (Fujian, China) and were used for mucus preparation as previously described.Citation31 After careful washing in PBS (0.01 mol/L pH 7.2), the skin mucus was collected by gently scraping the skin surface using a soft rubber spatula and was then homogenized in PBS. The homogenate was centrifuged twice (20 000g, 4 °C, 30 min), and the supernatant was filtered through 0.45-μM and 0.22-μM pore-size filters. Finally, PBS was added to each mucus sample to obtain a final concentration of 1 mg protein/mL, as determined by a Bradford assay.Citation32
In vitro adhesion assay
The bacterial adhesion assay was carried out as described in our previous study.Citation33 P. crocea mucus (50 μL) was evenly coated onto a 22-mm2 area of a glass slide area and fixed by methanol for 20 min. One milliliter of a bacterial suspension (108 colony-forming unit/mL) was spotted onto a mucus-coated glass slide. Then, the glass slides were placed in a humidified chamber, incubated at 25 °C for 2 h and then washed five times in PBS. Finally, the bacteria were fixed with 4% methanol for 30 min, stained with crystal violet for 3 min and counted under a microscope (× 1000). Three independent biological replicates (three technical replicates within each) were performed per group, and 20 random fields within each technical replicate were chosen to count. For the transient silencing, wild V. alginolyticus containing negative control siRNA (non-targeting) was used as a control group. For the stable silencing, wild V. alginolyticus containing empty plasmid pACYC184 was used as a control group.
RNA extraction and reverse transcription
Total bacterial RNA was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. First-strand complimentary DNA (cDNA) synthesis was performed with a RevertAid Mu-MLV cDNA Synthesis Kit (TransGen Biotech) following the manufacturer’s instructions.
qRT-PCR
The expression levels of genes were determined by qRT-PCR using QuantStudio 6 Flex (Life Technologies, Grand Island, NY, USA). Expression levels were normalized to 16S rRNA and then calculated using the 2−ΔΔCt method; primer sequences are listed in Supplementary Table S3.
Transmission electron microscope observation
Wild-type and stably silenced V. alginolyticus were cultured in LB20 broth at 28 °C for 16 h. Then, the culture was mounted onto formvar-coated copper grids by floating the grids on drops of sample for 1 min. The grids were washed three times in sterile deionized water to remove the medium. The cells were negatively stained by floating the grids on a drop of 1% phosphotungstic acid and then viewed using a transmission electron microscope (Philips, Tecnai F20, Amsterdam, Holland). More than 20 fields of view were randomly selected in each group. The length and diameter of flagella were measured with the Osiris 4.0 software (Geneva, Switzerland) from images (n=6 cells per condition).
Soft agar plate motility assay
The soft agar method was used to assay the motility of V. alginolyticus strains. Briefly, fresh overnight cultures were diluted to an OD660 of 0.03 in LB20. The cell suspension (1 μL/drop) was dropped onto the center of semisolid agar plates (LB20 broth+0.5% agar) and the plates were allowed to incubate for 20 h at 28 °C before measuring the colony diameters; three independent biological replicates (three technical replicates within each) were performed per group.
Biofilm assay
The biofilm assay for V. alginolyticus was implemented based on Lee et al.Citation34 V. alginolyticus strains were grown (overnight, 28 °C) in LB20 and then adjusted to the same OD600. Bacterial culture (50 μL) was added to 150 μL of LB20 per well of a 96-well plate and then incubated at 28 °C for 24 h. Wells were rinsed with sterile PBS, incubated (30 min) with 200 μL crystal violet (1%), washed with sterile PBS and air-dried. The stained biofilm was solubilized with 200 μL 33% acetic acid and then quantitated by measuring OD570; three independent biological replicates (three technical replicates within each) were performed per group.
Quantification of exopolysaccharide
Purification and quantification of exopolysaccharide (EPS) were performed using a slight modification of the methods of Acosta et al.Citation35 and Kimmel et al.,Citation36 respectively. Overnight cultures of V. alginolyticus were boiled (10 min) and centrifuged (12 000 rpm, 20 min) to remove cells. Trichloroacetic acid (10% final concentration) was added to precipitate proteins and polypeptides. After centrifugation, a filter membrane (0.45 μM) was used to filter the supernatant. Ice-cold ethanol (1 volume) was added to the supernatant fluid, and the coarse EPS was precipitated. After centrifugation, it was re-suspended in sterile distilled water (1/10th the original volume) and dialyzed in dialysis tubing. Phenol (0.3 mL, 6%) was added to the dialyzed EPS (0.5 mL), and then 1.5 mL concentrated sulfuric acid was quickly added and the mixture was shaken. This solution was heated for 15 min in a 100 °C water bath, cooled in cold water and finally quantitated by measuring OD490; three independent biological replicates (three technical replicates within each) were performed per group.
Hemolysis assay
Hemolysis assays were carried out as described by Tsou and Zhu.Citation37 Rabbit blood (Ping Rui Biotechnology Co., Ltd, Beijing, China) was prepared by washing three times with PBS. Washed rabbit blood (5 μL) was added to 245 μL of culture supernatants and incubated at 37 °C for 1 h under the relatively mild shaking conditions. After incubation, samples were centrifuged, and the released hemoglobin was measured by OD540. The percentage of total hemolysis was calculated by comparing the OD540 of the samples with positive (100% lysis by 1% Triton X-100) and negative controls.
Data processing
The data are presented as the mean±SD. Statistical analysis was performed by one-way analysis of variance with Dunnett’s test using the SPSS 13.0 software (Chicago, IL, USA). P-value<0.05 were considered statistically significant.
RESULTS
RNA-seq screening and validation for DEGs
The RNA-seq results for V. alginolyticus cultured under Cu2+, Pb2+, Hg2+ and low-pH conditions (V. alginolyticus cultured under normal condition was used as a control group) have been deposited in the NCBI Sequence Read Archive (Accession NO SRP049226). RNA-seq and DEG analysis finally yielded 1637, 1085, 846 and 1791 DEGs in the Cu-, Pb-, Hg- and low-pH-treated groups compared with the control group, respectively. Analysis of GO categories showed that the functional distribution of the DEGs from each stressed group was similar. In the libraries, most of the corresponding biological process genes were involved in cellular processes, metabolic processes, establishment of localization and localization. Most of the cellular component genes encoded proteins associated with cell, cell part, membrane and membrane part; moreover, most of the molecular function genes were associated with binding and catalytic activity. Using KEGG, DEGs were assigned to 164 KEGG pathways. Those pathways with the greatest representation by DEGs were ‘ABC transporter system’, ‘Two-component system’, ‘Glyoxylate and dicarboxylate metabolism’ and ‘Flagellar assembly’. These annotations provide a substantial resource for investigating specific processes, functions and pathways during bacteria adherence. Among the 164 KEGG pathways, some pathways are known to be closely related to adhesion, for example, ‘Two-component system’, ‘Bacterial chemotaxis’ and ‘Flagellar assembly’. In the present research, the Two-component system was selected for further research. GO and KEGG analyses revealed that there are 67 DEGs involved in Two-component systems. Among the DEGs, FlrA, FlrB and FlrC were generally significantly downregulated in all four of the stressed groups ().
To validate the results obtained by RNA-Seq, qRT-PCR was performed on the three genes. The qRT-PCR results were consistent with those in the RNA-Seq results. The Cu2+, Pb2+, Hg2+ and low-pH treatments significantly downregulated the expression of flrA (by 7.56-, 7.72-, 10.26- and 2.83-fold, respectively), flrB (by 8.27-, 7.61-, 11.22- and 2.60-fold, respectively) and flrC (by 9.55-, 5.60-, 6.17- and 2.65-fold, respectively; ). These data further reinforced the reliability of the RNA-Seq data in this study.
Figure 1 qRT-PCR analysis of the expression of flrA, flrB and flrC after stress treatments compared with untreated control. The data are presented as the means±SD; each treatment consisted of six independent biological replicates (three technical replicates within each). **P<0.01 compared with control subjects. qRT-PCR, quantitative reverse transcriptase real-time polymerase chain reaction.
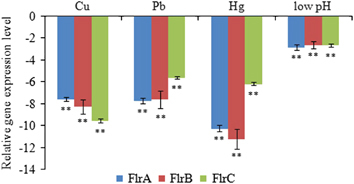
Table 1 Fold change of flrA, flrB and flrC identified by RNA-seq
Effects of transient gene silencing
The mRNA levels of the three target genes in V. alginolyticus were determined at 1, 6, 12 and 24 h after electrotransformation. Treatment of V. alginolyticus with siRNA () induced significant gene silencing (P<0.05) in V. alginolyticus show at 1–6 h; however, the silencing effect of each siRNA was not significant at any of the time points after 6 h. The decline in mRNA levels of each target gene revealed that the siRNA worked.
Figure 2 Transient gene silencing reduced the adhesion of V. alginolyticus. (A) qRT-PCR analysis of the expression of flrA, flrB and flrC after transient gene silencing at 1 h, 6 h, 12 h and 24 h compared with the control. The data are presented as the means±SD; each treatment consisted of six independent biological replicates (three technical replicates within each). *P<0.05 and **P<0.01 compared with control subjects. (B) The adhesion capacity to mucus of transient silenced V. alginolyticus at 2 h. The data are presented as the means±SD; three independent biological replicates (three technical replicates within each) were performed per group. *P<0.05 and **P<0.01 compared with control subjects. qRT-PCR, quantitative reverse transcriptase real-time polymerase chain reaction.
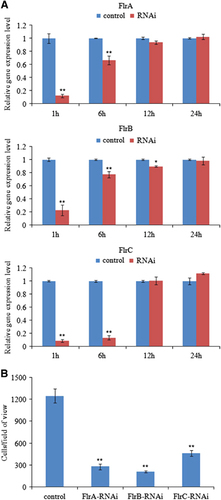
Thus, the in vitro assay for V. alginolyticus adhesion was performed under normal and RNAi conditions to further verify the silencing effect of siRNA at 2 h after electrotransformation. The number of adherent cells was counted. In the control group, ~1248±96 cells/view adhered to the glass. In contrast, in the flrA-, flrB- and flrC-RNAi groups, ~277±40, 207±12 and 463±38 cells/view adhered to the glass, respectively ().
These data show that the number of adherent cells was significantly lower in the RNAi groups than in the control group. The results of this adhesion assay indicate that RNAi significantly impairs V. alginolyticus adhesion to P. crocea mucus.
Effects of stable gene silencing
The mRNA levels of flrA, flrB and flrC were significantly lower (by 12.86-fold, 3.71-fold and 5.03-fold, respectively) in the stably silenced V. alginolyticus (). These data indicate that the stable gene silencing was reliable in this study.
Figure 3 Stable gene silencing reduced the adhesion of V. alginolyticus. (A) qRT-PCR analysis of the expression of flrA, flrB and flrC after stable gene silencing compared with the control. The data are presented as the means±SD; six independent biological replicates (three technical replicates within each) were performed per group. **P<0.01 compared with control subjects. (B) The adhesion capacity of stably silenced V. alginolyticus to mucus. The data are presented as the means±SD; three independent biological replicates (three technical replicates within each) were performed per group. **P<0.01 compared with control subjects. qRT-PCR, quantitative reverse transcriptase real-time polymerase chain reaction.
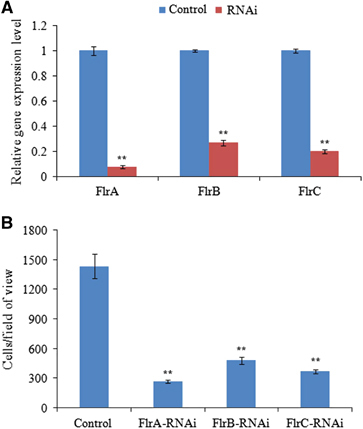
The stably silenced V. alginolyticus exhibited a significant decrease in adhesion. In the control V. alginolyticus, ~1435±122 cells/view adhered to the glass. However, in the flrA-, flrB- and flrC-RNAi V. alginolyticus, ~263±15, 480±36 and 367±18 cells/view adhered to the glass, respectively (). The adhesion assay results indicate that stable gene silencing significantly impairs V. alginolyticus adhesion to P. crocea mucus.
The expression levels of 12 important downstream virulence genes in the stably silenced V. alginolyticus are shown in . Compared with the control group (strain with the empty plasmid pACYC184), the expression levels of fliA, flgH, fliS, fliD, cheR, cheV and V12G01_22158 exhibited significant downregulation in all three of the RNAi groups, whereas the expression levels of toxT, ctxB, acfA, hlyA and tlh exhibited upregulation in the flrA- and flrC-RNAi groups. These data indicate that the expression levels of the majority of these virulence genes were affected by RNAi of flrA, flrB and flrC. For the upregulated genes, flrA-RNAi exhibited a more powerful effect than flrB-RNAi and flrC-RNAi. Furthermore, the expression of toxT, ctxB, acfA, hlyA and tlh was unchanged in the flrB stably silenced strain compared with the control. For the downregulated genes, flrB-RNAi exhibited a more powerful effect than flrA-RNAi and flrC-RNAi; however, the decrease in fliD transcription was most obvious in the flrA stably silenced strain.
Table 2 Expression of 12 important virulence genes after stable gene silencing
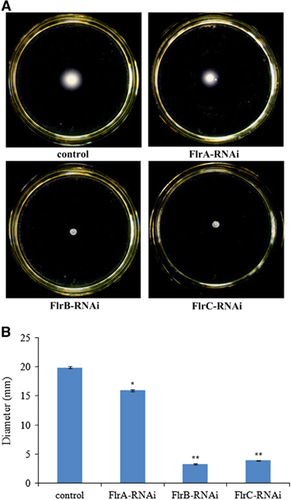
The flagella of stably silenced clones were observed (). The data revealed that flagella were present on the control and flrA-RNAi strains, whereas the flagella of the flrA-RNAi strain were significantly thinner and shorter compared with the control (P<0.01). However, no flagella could be observed on the flrB-RNAi and flrC-RNAi strains.
shows different levels of V. alginolyticus motility on soft agar plates. The results indicated that flrA-RNAi, flrB-RNAi and flrC-RNAi resulted in reduced motility of V. alginolyticus, whereas flrB-RNAi and flrC-RNAi were more effective than flrA-RNAi.
Figure 4 Detection of flagella in stably silenced V. alginolyticus strains. (A) Transmission electron micrographs of stably silenced V. alginolyticus strains. (B) Length and diameter of flagella in the control and flrA-RNAi strains. The data are presented as the means±SD; **P<0.01 compared with control subjects.
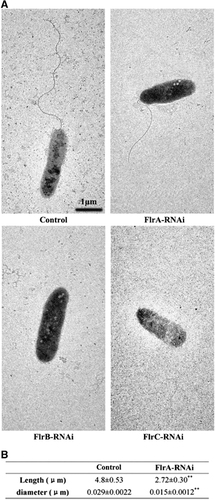
Figure 5 The motility behavior on soft agar plates of stably silenced V. alginolyticus strains. (A) Typical images of spreading of stably silenced V. alginolyticus strains and the control. (B) Diameter of the colony of each strain. The data are presented as the means±SD; three independent biological replicates (three technical replicates within each) were performed per group. *P<0.05 and **P<0.01 compared with control subjects.
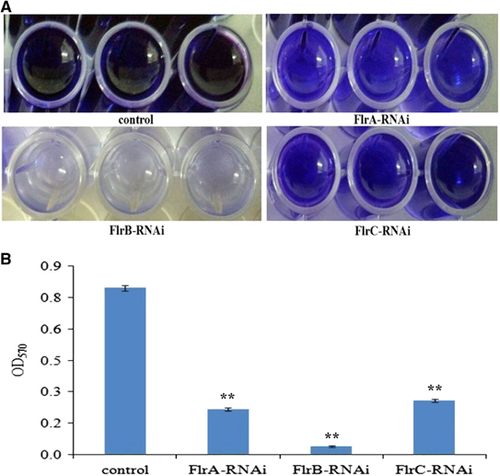
Biofilm formation of stably silenced clones was measured at OD570 (). The data showed that the ability to form biofilms was significantly decreased in flrA-RNAi, flrB-RNAi and flrC-RNAi compared to the wild-type strain, whereas flrB-RNAi was more effective than flrA-RNAi and flrC-RNAi (P<0.05).
Figure 6 Biofilm formation of stably silenced V. alginolyticus strains. (A) Typical images of stained biofilm of stably silenced V. alginolyticus strains and the control. (B) OD570 of stained biofilm in the colony of each strain. The data are presented as the means±SD; three independent biological replicates (three technical replicates within each) were performed per group. **P<0.01 compared with control subjects.
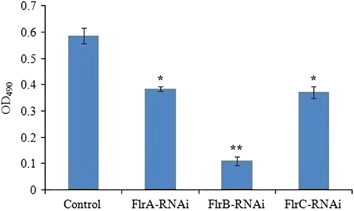
shows the production of EPS in different V. alginolyticus strains. The results revealed that EPS production of flrA-RNAi, flrB-RNAi and flrC-RNAi was significantly lower than the control, whereas the EPS production of flrB-RNAi was the lowest of the three stably silenced strains (P<0.05).
Figure 7 Production of EPS for stable silenced V. alginolyticus strains. The data are presented as the means±SD; three independent biological replicates (three technical replicates within each) were performed per group. *P<0.05 and **P<0.01 compared with control subjects. exopolysaccharide, EPS.
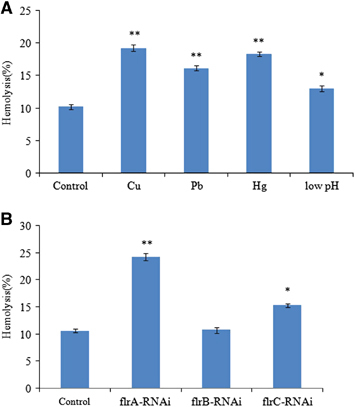
The hemolytic activity of V. alginolyticus strains during stress conditions of Cu2+, Pb2+, Hg2+ and low pH, and during stable gene silencing has been detected (). The V. alginolyticus strains under stress conditions of Cu2+, Pb2+, Hg2+ and low pH showed increased levels of a hemolytic activity against rabbit erythrocytes compared with the V. alginolyticus strains under normal condition (). The shows that hemolytic activity was significantly increased in flrA-RNAi and flrC-RNAi compared with the wild-type strain, and flrA-RNAi was more effective than flrC-RNAi, whereas hemolytic activity was not significantly increased in flrB-RNAi compared with the wild-type strain.
Figure 8 The hemolytic activity of V. alginolyticus strains under stress conditions of Cu2+, Pb2+, Hg2+ and low pH (A) and after stable gene silencing (B). The data are presented as the means±SD; three independent biological replicates (three technical replicates within each) were performed per group. *P<0.05 and **P<0.01 compared with control subjects.
Effects of different environmental conditions on the expression of flrA, flrB and flrC
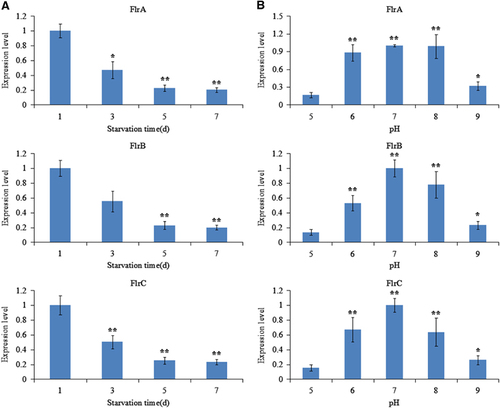
The expression of flrA, flrB and flrC under different environmental conditions was measured by qRT-PCR, and the results are shown in and .
The expression levels of flrA, flrB and flrC were essentially consistent under the different environmental conditions tested. flrA, flrB and flrC exhibited in part significant variation at different temperatures, with the highest expression levels of all the three genes occurring at 28 °C (). flrA, flrB and flrC expression levels were the lowest at 1.5% salinity; however, they later rose with increasing salinity (). Increasing starvation time also tended to reduce the expression levels of flrA, flrB and flrC (). Expression of flrA, flrB and flrC was significantly affected by pH, and the highest expression levels of all the three genes occurred at pH 7 (). These data indicate that flrA, flrB and flrC may participate in regulation of adhesion in the natural environment.
Figure 9 qRT-PCR analysis of the expression of flrA, flrB and flrC in the V. alginolyticus under different temperatures (A) and salinities (B). The data are presented as the means±SD; each treatment consisted of six independent biological replicates (three technical replicates within each). *P<0.05 and **P<0.01 compared with control subjects. qRT-PCR, quantitative reverse transcriptase real-time polymerase chain reaction.
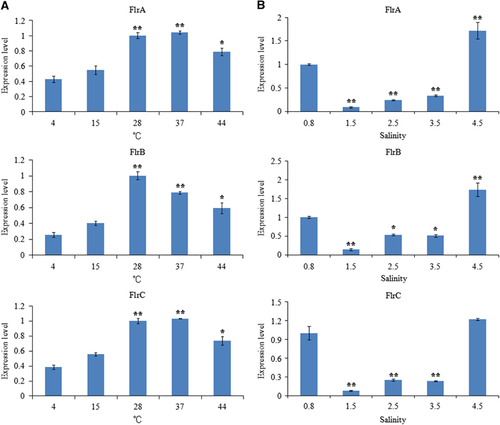
Figure 10 qRT-PCR analysis of the expression of flrA, flrB and flrC in the V. alginolyticus under different starvation time (A) and various pH values (B). The data are presented as the means±SD; each treatment consisted of six independent biological replicates (three technical replicates within each). *P<0.05 and **P<0.01 compared with control subjects. qRT-PCR, quantitative reverse transcriptase real-time polymerase chain reaction.
DISCUSSION
Bacterial adhesion is ultimately regulated by genes.Citation38 Several adhesion-related genes have been identified in V. alginolyticus,Citation3 Pseudomonas putida,Citation39 P. aeruginosa,Citation39 Candida albicans,Citation40 E. coliCitation41 and Erysipelothrix rhusiopathiae.Citation42 However, more details about the mechanism have not been found.
In our previous research,Citation14 V. alginolyticus adhesion was significantly reduced by Cu2+, Pb2+, Hg2+ and low pH. What’s more, flrA, flrB and flrC were generally downregulated under the four stress conditions tested. The flagellum has been shown to be involved in bacterial adhesion,Citation16 whereas flrA, flrB and flrC are required for the synthesis and assembly of the flagellum.Citation21 We do not think this is a coincidence. Therefore, we hypothesized a connection between them. In the present study, RNAi-mediated silencing of these three genes reduced bacterial adhesion. These results demonstrate that flrA, flrB and flrC have a key role in V. alginolyticus adhesion.
Bacterial adhesion has been demonstrated to be affected by bacterial motilityCitation19 and chemotaxis.Citation16 Indeed, bacterial motion and chemotaxis rely on the synthesis and assembly of the flagellum.Citation43 Transcription of the polar flagellar genes in V. cholerae has been categorized into four gene classes.Citation44 In V. cholerae, the sole class I gene identified so far is flrA, which encodes a σ54 (RpoN)-dependent transcriptional activator.Citation17 FlrA and RpoN-bound RNA polymerase could activate the transcription of the class II flagellar genes.Citation21 Of the Class II genes, fliA (σ28), flrB and flrC encode regulatory factors. flrB undergoes autophosphorylation and then activates flrC by transferring a phosphate to the conserved aspartate-54 (D54) residue at the amino terminus of flrC (flrC-P), allowing it to activate the σ54-dependent transcription of class III genes, fliA activates the transcription of class IV genes. Of the Class III genes, the flgH, cheR, fliD and fliS encode the L ring, methyltransferase, filament cap and flagellin chaperone, respectively.Citation45 Of class IV genes, cheV encodes the chemotaxis signal transduction protein.Citation17 fliA, flgH, fliS, fliD, cheR and cheV were dramatically decreased in the three stably silenced V. alginolyticus strains (). Nevertheless, it is hard to draw a common conclusion for the regulatory mechanism from the presented experimental results. Interestingly, the downregulation of fliA, flgH, fliS, cheR and cheV in the flrB-RNAi strain was more distinct than in the flrA-RNAi and flrC-RNAi strains. We also showed that the flrB-RNAi has more significant effects on flagella assembly and motility than the flrA-RNAi and flrC-RNAi strains. In this regard, flrB may have a major role in the regulation of flagellar synthesis and chemotaxis. However, the only previously known role the regulator flrB had was in flagellar stimulation and transcription of the chemotaxis gene. In addition, in the flrB-RNAi strain, downregulation of fliA was the most significant compared with flgH, fliS, fliD, cheR and cheV, suggesting that it has a vital role in the regulatory mechanism. As discussed above, the downregulation of fliA, flgH, fliS, fliD, cheR and cheV may be mainly concerned with the downregulation of transcription of flrA, flrB and flrC; and then the downregulation of fliA induced the downregulation of cheV. Thus, the six flagellar genes could be regulated by flrA, flrB and flrC. Downregulation of the expression of these genes may reduce the synthesis and assembly of the MS ring, filament, filament cap, motor/switch complex and chemotaxis protein. In the present study, the flagella of the flrA-RNAi strain were significantly thinner and shorter, whereas no flagella could be observed in the flrB-RNAi and flrC-RNAi strains (). The stably silenced strains were less motile compared with the control group (). These results were consistent with our speculation. Thus, flrA, flrB and flrC are very important and may affect bacterial adhesion via motility and chemotaxis.
The bacterial adhesion process was also directly mediated by adhesins,Citation16, Citation46 which are widely distributed on the bacterial surface.Citation47 V12G01_22158 is a gene that has recently been identified in pathogenic V. alginolyticus and encodes a GGDEF proteinCitation48 whose domain DGCs catalyze the conversion of two guanosine triphosphate into bis-(3′-5′)-cyclic dimeric guanosine monophosphate, which generally has an essential role as a ubiquitous second messenger in regulating motility, virulence, biofilm formation (associated with adhesins)Citation49 and EPS (an adhesin) biogenesis.Citation50 These results indicate that V12G01_22158 may be an important pathogenic gene in V. alginolyticus. Our data show that the expression of V12G01_22158 was significantly downregulated in every stably silenced V. alginolyticus strains (), and the three stably silenced strains had reduced biofilm formation and EPS production compared with the wild-type strain ( and ), especially in the flrB-RNAi strain. These results indicate that the transcription of V12G01_22158 was controlled by flrA, flrB and flrC. Thus, the downregulation of V12G01_22158 expression may be one of the reasons for the reduced adhesion of the stably silenced strains.
Genes acfA, ctxB, hlyA and tlh have a pivotal role in the colonization and pathogenesis of Vibrio. The hlyA and tlh are hemolysin genes.Citation45 toxT activates transcription of acfA and ctxB.Citation51 It has been shown that many virulence genes are more highly expressed in flagellar regulatory mutants and can enhance V. cholerae pathogenesis.Citation45 In this study, flrA-RNAi resulted in significant upregulation of toxT, acfA, ctxB, hlyA and tlh, which led to significant increase in hemolytic activity. flrB-RNAi exhibited no significant effect on the expression of these five genes and hemolytic activity. flrC-RNAi resulted in significant upregulation of toxT, ctxB and tlh, which led to significant increase in hemolytic activity. Enhanced hemolytic activity was also observed for the V. alginolyticus strains under stress conditions of Cu2+, Pb2+, Hg2+ and low pH, whereas flrA and flrC were downregulated by these conditions. Taken together, these results indicated that (i) transcriptions of acfA, ctxB, hlyA, tlh and toxT were negatively regulated by flrA, and then the hemolytic activity was repressed by flrA through hlyA and tlh; (ii) transcriptions of ctxB, tlh and toxT were negatively regulated by flrC, and then the hemolytic activity was repressed by flrC through tlh; (iii) the increase in acfA and ctxB expression is most likely because of the upregulation of toxT, which directly binds and activates acfA and ctxB.Citation45
Our results suggested that expression of the virulent genes toxT, ctxB and tlh was regulated by flrA and flrC, that the virulence genes acfA and hlyA were regulated by flrA, and that fliA, flgH, fliS, fliD, cheR, cheV and V12G01_22158 were regulated by flrA, flrB and flrC. flrA, flrB and flrC therefore have a significant signaling role in the pathogenicity of flagellation, motility, chemotaxis, adhesion, colonization and cytotoxicity. Moreover, flrB-RNAi led to maximum inhibition of genes associated with flagellation, motility, chemotaxis and adhesins, which translated to significant inhibition of adhesion; yet, flrB-RNAi did not increase the expression of genes associated with colonization or cytotoxicity. Therefore, flrB may be the most important gene among flrA, flrB and flrC from the perspective of disease prevention and treatment.
Many pathogenic bacteria can induce an adaptable response to environmental stimuli, primarily by altering gene expression.Citation19 For example, V. cholera rpoS mutants are sensitive to environmental conditions (including starvation, high osmolarity and oxidative stresses) when compared with the wild type.Citation52, Citation53 In our present study, the expression levels of flrA, flrB and flrC were found to be sensitive to different temperatures, changes in pH, changes in salinities and increased starvation time ( and ). Our previous research showed that V. alginolyticus adhesion was remarkably influenced by those environmental factors.Citation11 These results suggested that the changes in expression of flrA, flrB and flrC may be an important factor influencing adhesion under those environmental conditions and further strengthened the necessity of the present study.
In conclusion, our results suggest that (i) flrA, flrB and flrC are closely associated with the process of V. alginolyticus adhesion; (ii) flrA, flrB and flrC affect adhesion by affecting motility, chemotaxis and adhesins; and (iii) flrA, flrB and flrC regulate adhesion in different natural environments. In addition, our results showed that flrA, flrB and flrC have a significant signaling role in the pathogenicity of flagellation, motility, chemotaxis, adhesion, colonization and cytotoxicity. In this paper, we present for the first time the biochemical and molecular features of the flrA, flrB and flrC in V. alginolyticus and also indicate the pathogenic implications of these genes, which provide insight into prevention and treatment of this and related diseases.
Supplementary Table 1
Download PDF (6.7 KB)Supplementary Table S12
Download PDF (83.7 KB)Supplementary Table 3
Download PDF (170.1 KB)This work was supported by grants from Science and Technology Program of Xiamen Southern Oceanographic Center under contract NO 14CZP032HJ06, Regional Demonstration of Marine Economy Innovative Development Project under contract NO 14PYY050SF03 and 12PYY001SF08, The National Natural Science Foundation of China under contract NO 31272699 and 31502194, Fujian Provincial Department of Science and Technology under contract NO JA15289 and 2015R1036-4, The Natural Science Foundation of Fujian Province under contract NO 2016J05080. American Journal Experts (AJE) is thanked for language editing.
Supplementary Information for this article can be found on the Emerging Microbes & Infections website (http://www.nature.com/emi)
- WangYD,HuangSJ,ChouHNet al.Transcriptome analysis of the effect of Vibrio alginolyticus infection on the innate immunity-related complement pathway in Epinephelus coioides.BMC Genomics2014; 15:1102.
- WuC,ZhangD,KanMet al.The draft genome of the large yellow croaker reveals well-developed innate immunity.Nat Commun2014; 5:5227.
- WangL,HuangL,SuYet al.Involvement of the flagellar assembly pathway in Vibrio alginolyticus adhesion under environmental stresses.Front Cell Infect Microbiol2015; 5:59.
- Pizarro-CerdaJ,CossartP.Bacterial adhesion and entry into host cells.Cell2006; 124:715–727.
- PanX,YangY,ZhangJR.Molecular basis of host specificity in human pathogenic bacteria.Emerg Microbes Infect2014; 3:e23.
- BenhamedS,GuardiolaFA,MarsMet al.Pathogen bacteria adhesion to skin mucus of fishes.Vet Microbiol2014; 171:1–12.
- BergstromK,Kissoon-SinghV,GibsonDLet al.Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa.PLoS Pathog2010; 6:e1000902.
- LiuW,RenP,HeSet al.Comparison of adhesive gut bacteria composition, immunity, and disease resistance in juvenile hybrid tilapia fed two different Lactobacillus strains.Fish Shellfish Immunol2013; 35:54–62.
- JørgensenMG,NielsenJS,BoysenAet al.Small regulatory RNAs control the multi‐cellular adhesive lifestyle of Escherichia coli.Mol Microbiol2012; 84:36–50.
- TaghaviS,WuX,OuyangLet al.Transcriptional responses to sucrose mimic the plant-associated life style of the plant growth promoting endophyte Enterobacter sp. 638.PLoS One2015; 10:e0115455.
- YanQ,ChenQ,MaSet al.Characteristics of adherence of pathogenic Vibrio alginolyticus to the intestinal mucus of large yellow croaker (Pseudosciaena crocea.Aquaculture2007; 269:21–30.
- ChenQ,YanQ,WangKet al.Portal of entry for pathogenic Vibrio alginolyticus into large yellow croaker Pseudosciaena crocea, and characteristics of bacterial adhesion to mucus.Dis Aquat Organ2008; 80:181–188.
- ChenQ,YanQ,MaSet al.Adhesion of pathogenic Vibrio alginolyticus to the gill mucus of Pseudosciaena crocea.Acta Oceanol Sin2007; 26:101–109.
- KongW,HuangL,SuYet al.Investigation of possible molecular mechanisms underlying the regulation of adhesion in Vibrio alginolyticus with comparative transcriptome analysis.Antonie van Leeuwenhoek2015; 107:1197–1206.
- HuangL,HuJ,SuYet al.Identification and characterization of three Vibrio alginolyticus non-coding RNAs involved in adhesion, chemotaxis, and motility processes.Front Cell Infect Microbiol2015; 5:56.
- HaikoJ,Westerlund-WikströmB.The role of the bacterial flagellum in adhesion and virulence.Biology2013; 2:1242–1267.
- MoisiM,JenulC,ButlerSMet al.A novel regulatory protein involved in motility of Vibrio cholerae.J Bacteriol2009; 191:7027–7038.
- WöstenMM,WagenaarJA,van PuttenJP.The FlgS/FlgR two-component signal transduction system regulates the fla regulon in Campylobacter jejuni.J Biol Chem2004; 279:16214–16222.
- BeierD,GrossR.Regulation of bacterial virulence by two-component systems.Curr Opin Microbiol2006; 9:143–152.
- WangH,AyalaJC,BenitezJAet al.Interaction of the histone-like nucleoid structuring protein and the general stress response regulator RpoS at Vibrio cholerae promoters that regulate motility and hemagglutinin/protease expression.J Bacteriol2012; 194:1205–1215.
- KloseKE,MekalanosJJ.Distinct roles of an alternative sigma factor during both free‐swimming and colonizing phases of the Vibrio cholerae pathogenic cycle.Mol Microbiol1998; 28:501–520.
- WilhelmsM,MoleroR,ShawJGet al.Transcriptional hierarchy of Aeromonas hydrophila polar-flagellum genes.J Bacteriol2011; 193:5179–5190.
- SrivastavaD,HsiehML,KhataokarAet al.Cyclic di‐GMP inhibits Vibrio cholerae motility by repressing induction of transcription and inducing extracellular polysaccharide production.Mol Microbiol2013; 90:1262–1276.
- ShiM,GaoT,JuLet al.Effects of FlrBC on flagellar biosynthesis of Shewanella oneidensis.Mol Microbiol2014; 93:1269–1283.
- YanQP,SuWJ.YQ. [Studies on Vibriosis in caged-cultured Pseudosciaena crocea (Richardson)].J Jimei Univ (Nat Sci)2001; 6:191–196,Chinese.
- GarciaT,OttoK,KjellebergSet al.Growth of Vibrio anguillarum in salmon intestinal mucus.Appl Environ Microbiol1997; 63:1034–1039.
- YanQ,ZhaoM,WangXet al.Adhesion mechanisms of Vibrio fluvialis to skin mucus of Epinephelus awoara.Chin J Oceanol Limnol2010; 28:260–266.
- LancashireJF,TerryTD,BlackallPet al.Plasmid-encoded Tet B tetracycline resistance in Haemophilus parasuis.Antimicrob Agents Chemother2005; 49:1927–1931.
- DarsignyM,BabeuJ-P,SeidmanEGet al.Hepatocyte nuclear factor-4α promotes gut neoplasia in mice and protects against the production of reactive oxygen species.Cancer Res2010; 70:9423–9433.
- ChoiK-H,SchweizerHP.mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa.Nat Protoc2006; 1:153–161.
- HuangL,HuangL,YanQet al.The TCA pathway is an important player in the regulatory network governing Vibrio alginolyticus adhesion under adversity.Front Microbiol2016; 7:40.
- BradfordMM.A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding.Anal Biochem1976; 72:248–254.
- YanQ,ChenQ,MaSet al.Characteristics of adhesion of Vibrio alginolyticus to the skin mucus of Pseudosciaena crocea.Acta Oceanol Sin2005; 28:100–105.
- LeeHS,GuF,ChingSMet al.CdpA is a Burkholderia pseudomallei cyclic di-GMP phosphodiesterase involved in autoaggregation, flagellum synthesis, motility, biofilm formation, cell invasion, and cytotoxicity.Infect Immun2010; 78:1832–1840.
- AcostaMP,ValdmanE,LeiteSet al.Biosorption of copper by Paenibacillus polymyxa cells and their exopolysaccharide.World J Microbiol Biotechnol2005; 21:1157–1163.
- KimmelSA,RobertsRF,ZieglerGR.Optimization of exopolysaccharide production by Lactobacillus delbrueckii subsp. bulgaricus RR grown in a semidefined medium.Appl Environ MicrobioI1998; 64:659–664.
- TsouAM,ZhuJ.Quorum sensing negatively regulates hemolysin transcriptionally and posttranslationally in Vibrio cholerae.Infect Immun2010; 78:461–467.
- DunneWM.Bacterial adhesion: seen any good biofilms lately?Clin Microbiol Rev2002; 15:155–166.
- DuqueE,de la TorreJ,BernalPet al.Identification of reciprocal adhesion genes in pathogenic and non‐pathogenic Pseudomonas.Environ Microbiol2013; 15:36–48.
- PuriS,LaiWK,RizzoJMet al.Iron‐responsive chromatin remodelling and MAPK signalling enhance adhesion in Candida albicans.Mol Microbiol2014; 93:291–305.
- RuheZC,TownsleyL,WallaceABet al.CdiA promotes receptor‐independent intercellular adhesion.Mol Microbiol2015; 98:175–192.
- DingY,ZhuD,ZhangJet al.Virulence determinants, antimicrobial susceptibility, and molecular profiles of Erysipelothrix rhusiopathiae strains isolated from China.Emerg Microbes Infect2015; 4:e69.
- WangQ,SuzukiA,MaricondaSet al.Sensing wetness: a new role for the bacterial flagellum.EMBO J2005; 24:2034–2042.
- ProutyMG,CorreaNE,KloseKE.The novel σ54‐and σ28‐dependent flagellar gene transcription hierarchy of Vibrio cholerae.Mol Microbiol2001; 39:1595–1609.
- SyedKA,BeyhanS,CorreaNet al.The Vibrio cholerae flagellar regulatory hierarchy controls expression of virulence factors.J Bacteriol2009; 191:6555–6570.
- BoyleEC,FinlayBB.Bacterial pathogenesis: exploiting cellular adherence.Curr Opin Cell Biol2003; 15:633–639.
- KlemmP,VejborgRM,HancockV.Prevention of bacterial adhesion.Appl Microbiol Biotechnol2010; 88:451–459.
- ShengL,LvY,LiuQet al.Connecting type VI secretion, quorum sensing, and c-di-GMP production in fish pathogen Vibrio alginolyticus through phosphatase PppA.Vet Microbiol2013; 162:652–662.
- GerlachRG,HenselM.Protein secretion systems and adhesins: the molecular armory of Gram-negative pathogens.Int J Med Microbiol2007; 297:401–415.
- HenggeR.Principles of c-di-GMP signalling in bacteria.Nat Rev Microbiol2009; 7:263–273.
- ChildersBM,KloseKE.Regulation of virulence in Vibrio cholerae: the ToxR regulon.Future Microbiol2007; 2:335–344.
- NielsenAT,DolganovNA,OttoGet al.RpoS controls the Vibrio cholerae mucosal escape response.PLoS Pathog2006; 2:e109.
- SilvaAJ,Syed ZafarS,WeiliLet al.Role of the histone-like nucleoid structuring protein in the regulation of rpoS and RpoS-dependent genes in Vibrio cholerae.J Bacteriol2008; 190:7335–7345.
