Abstract
Emerging Microbes & Infections (2017) 6, e29; doi:10.1038/emi.2017.12; published online 10 May 2017
Dear Editor,
Carbapenems are considered as the last resort of antibiotic for the treatment of infection but many Gram-negative organisms have developed resistance to this antibiotic through loss or alteration of outer membrane porin protein OprD, over-expression of efflux pump, hyperproduction of an AmpC-type-β-lactamase and/or carbapenemase.Citation1 Carbapenemases are β-lactamases with catalytic efficiencies for carbapenem hydrolysis, including enzymes from Ambler’s classes A (extended spectrum β-lactamase), B (metallo-β-lacatamase, MBL) and D (Oxacillinases, OXA). The serine carbapenemases are derivatives of class A (e.g., IMI, KPC, GES) and class D enzymes (e.g., OXA-23, OXA-40, OXA-48) that hydrolyze carbapenems poorly but able to confer resistance. MBLs (e.g., IMP, VIM, GIM, SPM-l) can hydrolyze all β-lactams, including carbapenems (with the exception of aztreonam).Citation1, Citation2 Dutch imipenemase (DIM-1), a novel subclass 1 MBL can significantly hydrolyze broad-spectrum cephalosporins and carbapenems.Citation3 IMP and Verona integron-encoded metallo-β-lactamase (VIM) derivatives are widespread MBLsCitation4 and can be harbored within gene cassettes embedded into class 1 integron structureCitation3 as we know integrons can act as expression vector for the genes captured in the cassette.Citation5
Pseudomonas stutzeri is considered as environmental pseudomonad species and rarely cause nosocomial infection.Citation6 IMP and VIM derivatives have been identified in P. stutzeri,Citation7 but their integration in integron gene cassettee has not been reported. DIM-1 embedded in a class 1 integron located on a 70 kb plasmid has been reported originally in P. stutzeri,Citation3 however, subsequently identified in other species and locations.Citation8 In this study, we report the presence of VIM-2 MBL along with retrieval of associated gene cassette in clinical P. stutzeri isolate. A report is presented here concerning association of multidrug-resistant (MDR) clinical P. stutzeri 40D2 (initially named as 40/D/Mac2) isolated from the pus sample of a diabetic patient with a deep tissue foot infection (cellulitis) in a tertiary care hospital in Dhaka, Bangladesh.Citation9 Initially, 20 isolates were recovered among which three carbapenem resistant Pseudomonas isolates (40/D/Mac2, 40/D/Swab1, 40/D/CIP+Cefo1) were retrieved belonging to a single genotype, detected through amplified ribosomal DNA restriction analysis. Isolate 40D2 was selected for detailed investigation.
Isolate 40D2 was fast growing, colony appeared as pale, irregular shaped, wrinkled, gummy and flat in Nutrient Agar medium (Oxoid, UK) and microscopically Gram-negative and rod shaped bacterium. The 16S rRNA gene amplification, sequencing (GenBank accession number KT716345) and phylogenetic analysis revealed that the isolate was identified as P. stutzeri (accession number KP202687.1) with 100% sequence identity.
The antibiotic susceptibility was assessed by standard disc diffusion and microdilution methods,Citation10 which demonstrated that the isolated P. stutzeri 40D2 was MDR, being resistant to imipenem (MIC 256 mg/L), meropenem, doripenem, nitrofurantoin, ampicillin, oxacillin, gentamicin, trimethoprim, ciprofloxacin, levofloxacin, nalidixic acid, cefalexin, cefuroxime, cefotaxime, cefepime and surprisingly aztreonam. The isolate showed reduced susceptibility to azithromycin and chloramphenicol and was susceptible to amikacin, polymyxin B and colistin. Plasmid isolation was done by both Alkaline Lysis method Citation11 and Minipreps plasmid DNA Purification kit (Wizard Plus SV, Promega, Madison, WI, USA). Both methods were able to retrieve plasmids of control strain Escherichia coli V517 (plasmid size 2.2–55.5 kb); however no plasmid of these size range was retrieved from P. stutzeri 40D2 under our experimental condition. Carbapenemase production was confirmed by Blue-Carba Test,Citation12 and MBL production was determined by combined disc test method.Citation13 The occurrence of MBLs (blaVIM-2-like, blaNDM-1-like, blaIMP-3-like, blaIMP-4 like) was analyzed by PCR (Supplementary Table S1) revealing the presence of blaVIM-2 gene and the amplicon sequencing was carried out in order to determine 100% nucleotide sequence identity to that of Pseudomanas aeruginosa strain 7052 integron (GenBank accession number AY943084).
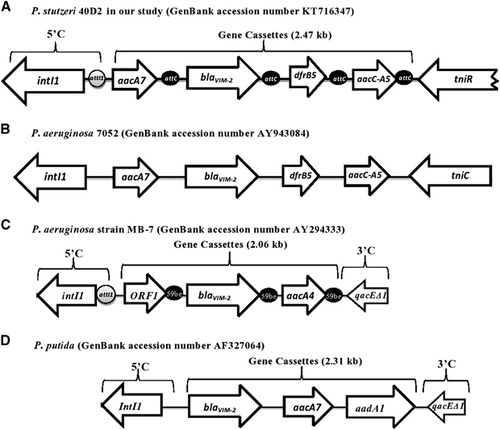
Integron class 1 associated gene cassettes were amplified using a total of 10 sets of primers (reference and designed primers, Supplementary Table S1). The amplicons were sequenced and the overlapping sequences were assembled to 4409 nucleotides (, GenBank accession number KT716347). After sequencing and homology analysis of the nucleotide sequences (http://www.ncbi.nlm.nih.gov), the location of blaVIM-2 gene cassette of P. stutzeri 40D2 was confirmed within class 1 integron. The cassettes include complete CDS of 1014 bp intI1 gene, 459 bp aacA7 gene, 801 bp blaVIM-2 gene, 237 bp dfrB5 gene, 477 bp aacC-A5 gene and 575 bp tniR (partial). The genome mapping (4409 bp) confirmed the presence and organization of MBL specific blaVIM-2 gene between aacA7 and dfrB5 gene (Figure ) comparable to the VIM-2 cassette of P. aeruginosa 7052; however, distinct from that of other Pseudomonas spp. (Figure ). The studied class 1 integron structure of 4409 bp, is named as In559 by INTEGRALL (http://integrall.bio.ua.pt/?acc=KT716347). The location of this CDS in chromosomal DNA has been identified through whole genome sequencing of 40D2 (accession number: MWUI00000000; contig 32 and 88). The aacA7 gene encodes aminoglycoside acetyltransferase, which confers resistance to aminoglycosides, but in our study P. stutzeri 40D2 was susceptible towards amikacin, an aminoglycoside. aacA7 gene encoded enzyme belongs to AAC (6’)-I subfamily Citation14 which is significant for amikacin resistance in P. aeruginosa; however, variants of this enzyme fail to provide amikacin resistance in clinical isolates.Citation15, Citation16 The unusual amikacin sensitivity in P. stutzeri 40D2 might be due to the presence of enzyme variants or accumulation of amikacin in bacterial cell due to alteration of OprD or efflux pump or other unknown mechanism that needs to be assessed further. Gentamicin resistance marker aacC-A5 encoding gentamicin 3'-acetyltransferase was found immediately downstream of dfrB5-encoding dihydrofolate reductase, which confers resistance to trimethoprim.
In summary, we present the first known worldwide report of the chromosomal located class 1 integron containing blaVIM-2 carbapenemase gene cassette embedded in tniR in P. stutzeri. Moreover, this is also the first known report of a VIM-2 producing P. stutzeri in Bangladesh. We further commence to perform complete genome sequencing of the retrieved isolate to reveal the inheritance of other mobile genetic elements and resistant gene markers. This investigation concludes that emergence of intI1 associated blaVIM-2 gene cassette-mediated carbapenem resistance in opportunistic pathogen P. stutzeri demands a serious public health concern because this gene cassette is considered as a xenogenetic pollutant causing acquisition of foreign resistant gene mostly in Gram-negative bacteria.
Ethical approval
The patient was aware of his involvement in this study and gave his consent to participate in this research work. This study was reviewed by ethical committee of faculty of Biological Science, University of Dhaka, Bangladesh.
Supplementary Table S1
Download PDF (78.5 KB)Acknowledgments
We thank TWAS (The World Academy of Science) for their gratuitous funding (Grant NO 15-123 RG/BIO/AS_I) during the research work. The present study was partially supported by the Ministry of Science and Technology, Government of Bangladesh by providing NST fellowship to Sabrin Bashar for her research work. We thank HEQEP (Higher Education Quality Enhancement Project), UGC (University Grants Commission) and Ministry of Education, Govt. of Bangladesh for equipment support. Also, we thank Mousumi Karmaker (the lecture of the department of Microbiology, Bangladesh University of Health Sciences) for isolation of the reported bacterium.
Supplementary Information for this article can be found on the Emerging Microbes & Infections website (http://www.nature.com/emi)
References
- Walsh TR, Toleman MA, Poirel Let al.Metallo-β-lactamases: the quiet before the storm? Clin Microbiol Rev 2005;18: 306–325.
- Queenan AM, Bush K.Carbapenemases: the versatile β-lactamases. Clin Microbiol Rev 2007;20: 440–458.
- Poirel L, Rodríguez-Martínez J-M, Al Naiemi Net al.Characterization of DIM-1, an integron-encoded metallo-β-lactamase from a Pseudomonas stutzeri clinical isolate in the Netherlands. Antimicrob Agents Chemother 2010;54: 2420–2424.
- Castanheira M, Bell JM, Turnidge JDet al.Carbapenem resistance among Pseudomonas aeruginosa strains from India: evidence for nationwide endemicity of multiple metallo-beta-lactamase clones (VIM-2, -5, -6, and -11 and the newly characterized VIM-18). Antimicrob Agents Chemother 2009;53: 1225–1227.
- Hall RM.Integrons and gene cassettes: hotspots of diversity in bacterial genomes. Ann N Y Acad Sci 2012;1267: 71–78.
- Ana PA, Gomes MZ, Silva ARet al.IMP-16 in Pseudomonas putida and Pseudomonas stutzeri: potential reservoirs of multidrug resistance. J Med Microbiol 2010;59: 1130–1131.
- Yan J-J, Hsueh P-R, Ko W-Cet al.Metallo-β-Lactamases in clinical Pseudomonas Isolates in Taiwan and identification of VIM-3, a novel variant of the VIM-2 enzyme. Antimicrob Agents Chemother 2001;45: 2224–2228.
- Booth MP, Kosmopoulou M, Poirel Let al.Crystal Structure of DIM-1, an acquired subclass B1 Metallo-β-Lactamase from Pseudomonas stutzeri. PloS One 2015;10: e0140059.
- Karmaker M, Sanyal SK, Sultana Met al.Association of bacteria in diabetic and non-diabetic foot infection- an investigation in patients from Bangladesh. J Infect Public Health 2015;9: 267–277.
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Fifteenth Informational Supplement M100-S15.Wayne, PA, USA: CLSI, 2005.
- Bimboim H, Doly J.A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res 1979;7: 1513–1523.
- Pires J, Novais A, Peixe L.Blue-Carba, an easy biochemical test for detection of diverse carbapenemase producers directly from bacterial cultures. J Clin Microbiol 2013;51: 4281–4283.
- Tsakris A, Poulou A, Pournaras Set al.A simple phenotypic method for the differentiation of metallo-β-lactamases and class A KPC carbapenemases in Enterobacteriaceae clinical isolates. J Antimicrob Chemother 2010;65: 1664–1671.
- Vanhoof R, Hannecart-Pokorni E, Content J.Nomenclature of genes encoding aminoglycoside-modifying enzymes. Antimicrob Agents Chemother 1998;42: 483.
- Galimand M, Lambert T, Gerbaud Get al.Characterization of the aac (6')-Ib gene encoding an aminoglycoside 6'-N-acetyltransferase in Pseudomonas aeruginosa BM2656. Antimicrob Agents Chemother 1993;37: 1456–1462.
- Poole K.Aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 2005;49: 479–487.