Abstract
Listeria is ubiquitous in a variety of environments and can be isolated from a wide range of animal hosts. Rodents are capable of carrying pathogenic bacteria in their intestines, such as Listeria, and can disseminate those pathogens into the natural environment and to where human activity occurs. In this study, we investigated the occurrence and antimicrobial susceptibility of Listeria spp. isolated from wild rodents found in natural environments in China. We collected 341 intestinal fecal samples of rodents from five different regions of China, all representing different rodent habitats. The antimicrobial susceptibility of the Listeria spp. isolates obtained were firstly assessed using the Kirby–Bauer disk diffusion method. Thirty-one samples were positive for Listeria spp., of which 11 were positive for Listeria monocytogenes and seven were positive for Listeria ivanovii. Other species identified include Listeria innocua, Listeria fleischmannii and Listeria floridensis. All Listeria spp. isolates were sensitive to the majority of the antimicrobials tested, but largely resistant to oxacillin (94.1%) and cefuroxime (70.6%). All L. monocytogenes isolates were further characterized by serotyping, multi-locus sequence typing (MLST) and pulsed-field gel electrophoresis (PFGE). L. monocytogenes strains were grouped into three serotypes, five sequence types and five pulsotypes (PTs) by serotyping, MLST and PFGE, respectively. Almost half of the isolates (five of 11) belonged to serotype 1/2b, ST87 and PT1. This study determined that Listeria is carried in the intestinal tracts of wild rodents from multiple regions at a low rate, filling an epidemiological data gap on Listeria in natural environments in China.
Emerging Microbes & Infections (2017) 6, e44; doi:http://10.1038/emi.2017.28; published online 7 June 2017
Introduction
Until recently the genus Listeria was thought to consist of six species, including L. monocytogenes, L. innocua, L. welshimeri, L. ivanovii, L. seeligeri and L. grayi. However, another 11 novel species have been reported since 2009: L. marthii, L. fleischmannii, L. floridensis, L. rocourtiae, L. weihenstephanensis, L. cornellensis, L. aquatica, L. riparia, L. grandensis, L. booriae and L. newyorkensis.Citation1, Citation2, Citation3, Citation4, Citation5, Citation6, Citation7 Only two species, L. monocytogenes and L. ivanovii, are pathogenic due to their respective species-specific virulence determinants. L. monocytogenes can cause severe invasive infections in both humans and animals, while L. ivanovii rarely causes infections in humans but is an important cause of infection in other animals, particularly in ruminants.Citation8, Citation9 Human infections by other Listeria species, such as L. seeligeri and L. innocua, are rare and are seen mainly in immunocompromised individuals.Citation10, Citation11 Human listeriosis, which is mainly caused by the consumption of L. monocytogenes in contaminated food products, has low morbidity but high mortality rates. The symptoms of listeriosis in humans are non-specific, varying from mild to severe illness. Immunocompetent adults may suffer a self-limiting febrile gastroenteritis.Citation12 In contrast, immunocompromised individuals, including people with severe underlying disease conditions, the elderly and newborns, may suffer septicemia and central nervous system infections.Citation13 Pregnant women may suffer miscarriage, preterm delivery or stillbirth, although only flu-like symptoms are manifested.Citation14
Listeria has been shown to be ubiquitously distributed in a variety of environments due to its adaptability. For example, Listeria can survive at a broad range of pH (4.5–9.2), temperature (0–45 °C) and salt concentrations (up to 10% NaCl).Citation15 Most Listeria spp. isolates are susceptible to many antimicrobials, except for some modern cephalosporins, dalfopristin/quinupristin, pipemidic acid, oxacillin and aztreonam.Citation16 The occurrence and antimicrobial resistance of Listeria from various food products and food-processing environments have been well studied.Citation17, Citation18 Recently, Listeria spp. isolated from food products or food-processing environments which are resistant to multiple antimicrobial agents have been reported worldwide.Citation19, Citation20 However, there is a lack of published information on the occurrence and antimicrobial resistance of Listeria spp. in wild animals from natural environments. In these environments, rodents could represent a reservoir for many pathogens. Shedding of Listeria spp., especially L. monocytogenes and L. ivanovii, in the feces of rodents could contaminate food products or food-processing environments by direct or indirect transmission paths. The present study was undertaken to determine the occurrence and antimicrobial resistance of Listeria spp. in rodents from natural environments.
L. monocytogenes isolates are classified into four lineages by a large number of subtyping methods.Citation21, Citation22, Citation23 The majority of L. monocytogenes isolates belong to lineage I (including serotype 1/2b, 3b, 4b, 4d and 4e) or lineage II (including 1/2a, 3a, 1/2c and 3c). A number of studies have shown that the majority of human listeriosis cases were associated with lineage I isolates, while major L. monocytogenes contaminations of food were associated with lineage II isolates. L. monocytogenes isolates assigned to lineage III or lineage IV, including serotype 4a and 4c, are rare and mostly isolated from ruminants.Citation24 Pulsed-field gel electrophoresis (PFGE) and multi-locus sequence typing (MLST) have been widely used for the epidemiological investigation of L. monocytogenes and source tracking of specific strains in outbreaks. Further molecular characterization for L. monocytogenes isolates from rodents was performed to provide insight on epidemiological features of this foodborne pathogen from natural environments.
MATERIALS AND METHODS
Sample collection
According to the medical research regulations of the Ministry of Health China, the present study was approved by the ethics committee of the National Institute for Communicable Disease Control and Prevention, China CDC (Approval NO. ICDC2014003). In this study, 341 intestinal fecal samples of rodents were collected from five regions (Tibet, Hainan, Guangdong, Fujian and Shanxi province) in China during September 2014–June 2015. The sampling areas comprised five different habitat types, including the junction area of farm and woodland, woodland, cassava field, grassland–shrubland and waste-grassland (Table ). The captured rodents were autopsied, and ~1 g of fecal content from the cecum and colon were collected in 5 mL of Brain Heart Infusion Broth (BHIB) containing 15% glycerol. The samples were stored at 4 °C for ~two weeks prior to experiments.
Table 1 Combined data on fecal samples of rodents collected and incidence of Listeria spp.
Isolation, identification and confirmation of Listeria spp.
Listeria strains were isolated according to the ISO 11290 method with modifications. The intestinal feces were introduced into 10 mL Half-Fraser broth and incubated at 30 °C for 24 h. Subsequently, 0.5 mL of the primary enrichment cultures were transferred to 4.5 mL Fraser broth and incubated at 37 °C for 48 h. A loopful of secondary enrichment was streaked onto Chromogenic Listeria Agar (Oxoid, Basingstoke, UK) and incubated at 37 °C for 24–48 h. After incubation, colonies suspected of being Listeria spp. based on color and morphology were selected for identification. Bacterial colonies were identified using 16S rDNA amplification and sequencing and the API Listeria test (bioMérieux, Marcyl’Etoile, France). All confirmed Listeria isolates were stored in BHIB containing 15% glycerol at −80 °C.
Antimicrobial susceptibility testing
Sensitivity of the Listeria isolates to 16 antimicrobials (10 μg gentamicin, 30 μg kanamycin, 10 μg streptomycin, 10 units penicillin G, 10 μg ampicillin, 1 μg oxacillin, 30 μg chloramphenicol, 5 μg rifampicin, 10 μg imipenem, 30 μg vancomycin, 2 μg clindamycin, 15 μg erythromycin, 30 μg tetracycline, 1.25/23.75 μg trimethoprim/sulfamethoxazole, 5 μg ciprofloxacin and 30 μg cefuroxime) was assessed using the disk diffusion technique according to the Clinical and Laboratory Standard Institute (CLSI). Briefly, pure frozen culture was transferred to BHIB and incubated at 37 °C overnight. A cell suspension was prepared by suspending colonies in 0.85% NaCl (w/v) until the turbidity was equal to 0.5 MacFarland standards; the suspension was spread onto the surface of Mueller–Hinton agar (Oxoid). The diameter of the inhibition zone surrounding each disk was measured after 18–24 h of incubation at 37 °C. The results for each antimicrobial were classified as sensitive, intermediate or resistant according to the CLSI criteria for Staphylococci spp.Citation25 Streptococcus pneumoniae ATCC 49619 was used as a control strain.
Serotyping, MLST and PFGE
L. monocytogenes serotype was identified by a combination of multiplex PCR and traditional slide agglutination. The multiplex PCR was performed by targeting the genes lmo0737, lmo1118, ORF2110, ORF2819 and Listeria-specific prs described by Doumith et al.Citation26 When the serogroups were identified, only the antiserum against somatic antigens was used. MLST based on seven house-keeping genes (abcZ, bglA, cat, dapE, dat, ldh and lhkA) was performed according to the method of Ragon et al.Citation27 The scheme and genotypic data are available at http://bigsdb.web.pasteur.fr/listeria/. Minimum spanning tree analysis was inferred using BioNumerics (Version 5.10, Applied Maths, Belgium). PFGE of the L. monocytogenes strains was performed using the primary restriction enzyme AscI according to the standard operating procedure by PulseNet of Centers for Disease Control and Prevention.Citation28 Similarities between the digestion profiles of strains were analyzed by unweighted pair group method with arithmetic mean using BioNumerics software (Version 5.10, Applied Maths).
Statistical analysis
X2 test or Fisher’s exact test, as appropriate, was performed in SAS 9.4 (SAS Institute Inc., Cary, NC, USA) to check the significant effect of the regions and the types of rodents on the occurrence of Listeria spp. A P-value <0.05 was considered as statistically significant. In order to reduce the sample collection bias, the rodents were categorized into six types based on genera, including Rattus (n=130), Niviventer (n=72), Apodemus (n=67), Mus (n=33), Bandicota (n=25), and others (including Phaiomys (n=9), Cricetulus (n=3) and Microtus (n=2)).
RESULTS
Occurrence of Listeria spp. in feces of rodents
Thirty-one out of 341 fecal samples were found to be positive for Listeria spp. (Table ). Three samples contained two Listeria species with two samples containing L. monocytogenes and L. innocua, and one sample containing L. monocytogenes and L. fleischmannii. The 34 Listeria spp. isolates included 11 L. monocytogenes (32.4%), seven L. ivanovii (20.6%), 10 L. innocua (29.4%), five L. fleischmannii (14.7%) and one L. floridensis (2.94%). By region and habitat, the prevalence rates of Listeria ranged from 5.3% (Guangdong) to 25.8% (Tibet) and from 3.2% (woodland) to 16.7% (grassland–shrubland; Table ). The incidence of Listeria in Tibet was significantly higher than in the other regions (P<0.05). The prevalence rates of Listeria in six types of rodents varied from 0% to 16% (Table ), however, the differences were not statistically significant (P>0.05).
Table 2 Summary data on the occurrence of Listeria spp. according to regions, habitat and species of rodents
Antimicrobial resistance of Listeria spp. strains
All the Listeria spp. isolates were tested for antimicrobial susceptibility (Table ). The most frequent antibiotic resistance was to oxacillin (94.1%), followed by cefuroxime (70.6%), clindamycin (20.6%) and tetracycline (11.8%). All isolates were sensitive to gentamicin, kanamycin, penicillin G, ampicillin, imipenem and vancomycin. All of the L. fleischmannii isolates were resistant to clindamycin. Moreover, one of the L. innocua isolates was found to be resistant to seven antibiotics tested, including streptomycin, oxacillin, chloramphenicol, clindamycin, trimethoprim/sulfamehtoxazole, tetracycline and cefuroxime.
Table 3 Antimicrobial resistance profile of Listeria spp. isolates tested in this study
Genotypic characterization of L. monocytogenes isolates
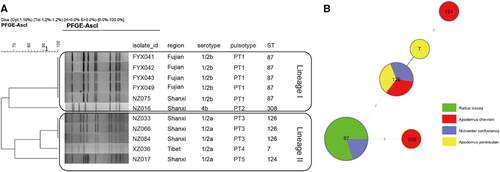
The 11 L. monocytogenes isolates were typed by serotyping, MLST and PFGE. Five isolates were serotype 1/2a, five were 1/2b and one was 4b. By MLST, the 11 isolates were divided into five sequence types (STs). There were five ST87, three ST126 and one each of ST7, ST124 and ST308. Isolates with the same ST also shared the same pulsotype, for example, all ST87 isolates were PT1, while all ST126 isolates were PT3 (Figure ). Four of the five ST87/PT1 strains were isolated from waste-grassland in Fujian. The minimum spanning tree of the L. monocytogenes isolates determined that there was no correlation between STs of L. monocytogenes and the species of rodents, although the sample set was small (Figure ).
DISCUSSION
Although there have been many reports of contamination of food products and food-processing environments with L. monocytogenes and other Listeria spp., there have been limited studies on the occurrence and characteristics of Listeria spp. in wild animals from natural environments. In this study, we found that the incidence of Listeria spp. in the feces of rodents was 9.97%, with L. monocytogenes and L. ivanovii at 3.23% and 2.05%, respectively. Tibet had the highest incidence of Listeria and was statistically significantly different from the other regions (P<0.05), although the number of positive samples was small. It is interesting to note that L. ivanovii was only isolated from Tibet. The higher isolation rate in the Tibet plateau may be attributed to the activities of a variety of wildlife, particularly ruminants, such as yaks, sheep and deer. However, this is unlikely due to the altitude. Kristina et al. found that a higher L. ivanovii isolation rate occurred in wildlife reserve regions and sites near the habitats of wild and domestic ruminants; the altitudes of the study sites were <500 m, which are much lower than the Tibet plateau (above 3000 m).Citation29
The prevalence of L. monocytogenes in rodents from natural environments was lower than those reported in different food products in China (ranging 5.3%–20%).Citation30, Citation31, Citation32 Higher prevalence in the latter may be because food products and food-processing environments provide a very suitable environment for the proliferation of Listeria spp. However, a recent survey on various food products from three large cities in central China observed a lower prevalence of L. monocytogenes (2.3%).Citation33
There has of yet been no reports focusing on Listeria spp. prevalence in wild animals in China. For Listeria spp., higher prevalence levels from rodents were reported in Japan by Iida et al. and Inoue et al., with rates of 17.1% and 24.5%, respectively.Citation34, Citation35 In addition, higher isolation rates of Listeria spp. were found in wild birds according to the studies of Yoshida et al. and Hellstrom et al., with rates of 24.5% and 53%, respectively.Citation36, Citation37 In contrast, lower prevalence rates of Listeria spp. were reported in domestic animals (with the exception of pig at 12.2%), including cats (0%), chickens (4.7%), dogs (2%), and cattle (5.1%) by Iida et al.,Citation34 as well as in wild non-rodent mammals (6.1%) by Yoshida et al.Citation36 For L. monocytogenes, higher isolation rates have been reported in rodents (6.5% and 5.7%)Citation34, Citation35 and wild birds (36%) in Japan,Citation37 as well as in livestock (5.9%) and wild animals (3.7%) in Canada.Citation38 In contrast, lower isolation rates have occurred in domestic animals ranging from 0% to 1.92% in the study of Iida et al.Citation34 and in wild non-rodent mammal animals (0.96%) and birds (0.5%) in the report of Yoshida et al.Citation36 For L. ivanovii, the isolation rates reported in rodents (1.6%) and wild non-rodent mammal animals (0.16%) were lower than those reported in this study.Citation35, Citation36 However, few studies have reported the prevalence of this Listeria species. Generally, different isolation rates have occurred in different hosts from different regions with no apparent trends. Overall, although the prevalence of Listeria spp., especially L. monocytogenes and L. ivanovii, in wild rodents from natural environments in China was low, surveillance on pathogenic Listeria in wild animals from natural environments as a reservoir is important to public health.
Many published studies have determined that the rate of antimicrobial resistance in Listeria is relatively low but has been increasing.Citation39, Citation40, Citation41 Similarly, we determined that almost all of the Listeria isolates were susceptible to the antibiotics commonly used to treat human listeriosis. Only one isolate was found to be resistant to trimethoprim/sulfamethoxazole, an alternative drug for patients allergic to penicillin.Citation42 Also, all of the L. fleischmannii isolates and the one L. floridensis isolate were resistant to clindamycin; the vast majority of the isolates were resistant to cefuroxime. Oxacillin resistance has previously been associated with L. monocytogenes, L. innocua and L. welshimeri isolates from various sources, including food products, the environment, animals and humans.Citation17, Citation43 It is interesting to note that the L. fleischmannii strains studied in Bertsch et al.Citation3 harbored a transferable transposon that confers resistance to clindamycin. It would be interesting to determine if our L. fleischmannii isolates also carry the resistance transposon. In addition, intermediate resistance against clindamycin was observed in nearly half of L. monocytogenes isolates (45%) and most of the L. innocua isolates (8, 80%). All but one of the L. ivanovii isolates exhibited intermediate resistance to streptomycin. Among the five Listeria species in this study, L. innocua overall displayed the highest resistance. Notably, one L. innocua isolate was resistant to seven of the antibiotics tested. Gomez et al. reported that 43.1% and 13.9% L. innocua strains isolated from meat products and meat-processing environments were one or two antimicrobials resistance and multidrug resistance, respectively.Citation43 High resistance to oxacillin, clindamycin and cefuroxime, as reported previously, may be intrinsic. However, resistance and intermediate resistance to many other antibiotics were surprising as Listeria residing in wild rodents would not face selection pressure from antibiotic use. A continuous surveillance of emerging resistance in Listeria is important in combating antibiotic resistance in human infections as the resistant isolates from wild animals in natural environments may serve as reservoirs of resistance genes.
In this study, subtyping of L. monocytogenes isolates by serotyping, PFGE and MLST provided further insights into the molecular characterization of this pathogen found in rodents in natural environments. All of the 11 L. monocytogenes isolates were classified into two previously defined lineages (Figure ). Serotype 1/2b and 4b strains belong to lineage I, whereas serotype 1/2a strains belong to lineage II. The serotypes found in this study were all primarily associated with human listeriosis serotypes.Citation24 The high prevalence of serotype 1/2a in this study was consistent with the results of other studies from China reporting that serotype 1/2a is one of the predominant serotypes from food products and food-processing environments in China.Citation19, Citation44 Serotypes 4b and 1/2c had also been reported as common serotypes in food products and food-processing environments by different surveys,Citation19, Citation45 although only one 4b and no 1/2c L. monocytogenes isolates were identified in this study. By MLST, the predominant ST found was ST87 which was in more than one rodent species and region. ST87 is also the predominant ST which causes human infections.Citation46 However the other four STs found in this study were not commonly found in contaminated foods or human infections in China.Citation45, Citation46
L. monocytogenes strains were primarily isolated from two of the five regions: Fujian (four strains) and Shanxi (five strains). The L. monocytogenes strains displayed different distribution characteristics between these two regions. In Fujian, the L. monocytogenes strains which were isolated from Rattus lossea in waste-grassland, had identical subtypes, with serotype 1/2b, ST87 and PT1, suggesting that this subtype persisted in this region. In Shanxi, the L. monocytogenes strains which were isolated from different species of rodents in the junction area of farm and woodland, were differentiated into four subtypes, demonstrating more genetic diversity. More studies will be required to further assess the genetic diversity of L. monocytogenes in these different regions and in different rodent species.
In conclusion, this is the first investigation of the occurrence and characteristics of Listeria spp. in wild rodents from natural environments in China. Listeria was carried in the intestinal tracts of wild rodents generally at low frequencies from multiple regions in China. However, by strain characteristics, L. monocytogenes from wild rodents possesses a potential health risk as some of the same serotypes are most frequently isolated from human infections. Therefore, rodents could spread the pathogen in natural environments by the fecal–oral route, and also to human living environments, potentially leading to disease in humans. Listeria spp. which were resistant to antibiotics were isolated in this study. There is a rising threat attributed to the transfer of antimicrobial resistance from non-pathogenic Listeria to pathogenic Listeria species. This study fills an epidemiological gap on Listeria in natural environments in China.
Acknowledgments
This work was funded by the Ministry of Science and Technology of China (Mega Project of Research on the Prevention and Control of HIV/AIDS, Viral Hepatitis Infectious Diseases 2011ZX10004-001, 2013ZX10004-101 to Changyun Ye and 2012ZX10004215) and State Key Laboratory of Infectious Disease Prevention and Control (2015SKLID507 to Changyun Ye). National Institute for Communicable Disease Control and Prevention, China CDC (2016ZZKTB09 to Changyun Ye).
References
- Graves LM, Helsel LO, Steigerwalt AGet al.Listeria marthii sp. nov., isolated from the natural environment, Finger Lakes National Forest. Int J Syst Evol Microbiol 2010;60: 1280–1288.
- Leclercq A, Clermont D, Bizet Cet al.Listeria rocourtiae sp. nov. Int J Syst Evol Microbiol 2010;60: 2210–2214.
- Bertsch D, Rau J, Eugster MRet al.Listeria fleischmannii sp. nov., isolated from cheese. Int J Syst Evolut Microbiol 2013;63: 526–532.
- den Bakker HC, Manuel CS, Fortes EDet al.Genome sequencing identifies Listeria fleischmannii subsp. coloradonensis subsp. nov., isolated from a ranch. Int J Syst Evol Microbiol 2013;63: 3257–3268.
- Lang Halter E, Neuhaus K, Scherer S.Listeria weihenstephanensis sp. nov., isolated from the water plant Lemna trisulca taken from a freshwater pond. Int J Syst Evol Microbiol 2013;63: 641–647.
- den Bakker HC, Warchocki S, Wright EMet al.Listeria floridensis sp. nov., Listeria aquatica sp. nov., Listeria cornellensis sp. nov., Listeria riparia sp. nov. and Listeria grandensis sp. nov., from agricultural and natural environments. Int J Syst Evol Microbiol 2014;64: 1882–1889.
- Weller D, Andrus A, Wiedmann Met al.Listeria booriae sp. nov. and Listeria newyorkensis sp. nov., from food processing environments in the USA. Int J Syst Evol Microbiol 2015;65: 286–292.
- Lessing MP, Curtis GD, Bowler IC.Listeria ivanovii infection. J Infect 1994;29: 230–231.
- Beye M, Gouriet F, Michelle Cet al.Genome analysis of Listeria ivanovii strain G770 that caused a deadly aortic prosthesis infection. New Microbes New Infect 2016;10: 87–92.
- Rocourt J, Hof H, Schrettenbrunner Aet al.Acute purulent Listeria seelingeri meningitis in an immunocompetent adult. Schweiz Med Wochenschr 1986;116: 248–251.
- Perrin M, Bemer M, Delamare C.Fatal case of Listeria innocua bacteremia. J Clin Microbiol 2003;41: 5308–5309.
- Sim J, Hood D, Finnie Let al.Series of incidents of Listeria monocytogenes non-invasive febrile gastroenteritis involving ready-to-eat meats. Lett Appl Microbiol 2002;35: 409–413.
- Doganay M.Listeriosis: clinical presentation. FEMS Immunol Med Microbiol 2003;35: 173–175.
- Delgado AR.Listeriosis in pregnancy. J Midwifery Womens Health 2008;53: 255–259.
- Hain T, Chatterjee SS, Ghai Ret al.Pathogenomics of Listeria spp. I J Med Microbiol 2007;297: 541–557.
- Troxler R, von Graevenitz A, Funke Get al.Natural antibiotic susceptibility of Listeria species: L. grayi, L. innocua, L. ivanovii, L. monocytogenes, L. seeligeri and L. welshimeri strains. Clin Microbiol Infect 2000;6: 525–535.
- Khen BK, Lynch OA, Carroll Jet al.Occurrence, antibiotic resistance and molecular characterization of Listeria monocytogenes in the beef chain in the Republic of Ireland. Zoonoses and Public Health 2015;62: 11–17.
- Camargo AC, de Castilho NP, da Silva DAet al.Antibiotic resistance of Listeria monocytogenes isolated from meat-processing environments, beef products, and clinical cases in Brazil. Microb Drug Resist 2015;21: 458–462.
- Wu S, Wu Q, Zhang Jet al.Prevalence, antibiotic resistance and genetic diversity of Listeria monocytogenes isolated from retail ready-to-eat foods in China. Food Control 2015;47: 340–347.
- Garedew L, Taddese A, Biru Tet al.Prevalence and antimicrobial susceptibility profile of listeria species from ready-to-eat foods of animal origin in Gondar Town, Ethiopia. BMC Microbiol 2015;15: 100.
- Rasmussen OF, Skouboe P, Dons Let al.Listeria monocytogenes exists in at least three evolutionary lines: evidence from flagellin, invasive associated protein and listeriolysin O genes. Microbiology 1995;141: 2053–2061.
- Wiedmann M, Bruce JL, Keating Cet al.Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect Immun 1997;65: 2707–2716.
- Ward TJ, Ducey TF, Usgaard Tet al.Multilocus genotyping assays for single nucleotide polymorphism-based subtyping of Listeria monocytogenes isolates. Appl Environ Microbiol 2008;74: 7629–7642.
- Orsi RH, den Bakker HC, Wiedmann M.Listeria monocytogenes lineages: genomics, evolution, ecology, and phenotypic characteristics. Int J Med Microbiol 2011;301: 79–96.
- Cockerill FR. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-First Informational Supplement, M100S21.Clinical and Laboratory Standards Institute (CLSI).2011.
- Doumith M, Buchrieser C, Glaser Pet al.Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J Clin Microbiol 2004;42: 3819–3822.
- Ragon M, Wirth T, Hollandt Fet al.A new perspective on Listeria monocytogenes evolution. PLoS Pathog 2008;4: e1000146.
- Graves LM, Swaminathan B.PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresis. Int J Food Microbiol 2001;65: 55–62.
- Linke K, Ruckerl I, Brugger Ket al.Reservoirs of listeria species in three environmental ecosystems. Appl Environ Microbiol 2014;80: 5583–5592.
- Wang G, Qian W, Zhang Xet al.Prevalence, genetic diversity and antimicrobial resistance of Listeria monocytogenes isolated from ready-to-eat meat products in Nanjing, China. Food Control 2015;50: 202–208.
- Wang W, Zhou X, Suo Yet al.Prevalence, serotype diversity, biofilm-forming ability and eradication of Listeria monocytogenes isolated from diverse foods in Shanghai, China. Food Control 2017;73(Part B): 1068–1073.
- Wu S, Wu Q, Zhang Jet al.Listeria monocytogenes prevalence and characteristics in retail raw foods in China. PLoS One 2015;10: e0136682.
- Du X-J, Zhang X, Wang X-Yet al.Isolation and characterization of Listeria monocytogenes in Chinese food obtained from the central area of China. Food Control 2017;74: 9–16.
- Iida T, Kanzaki M, Maruyama Tet al.Prevalence of Listeria monocytogenes in intestinal contents of healthy animals in Japan. J Vet Med Sci 1991;53: 873–875.
- Inoue S, Tanikawa T, Kawaguchi Jet al.Prevalence of Listeria (spp.) in wild rats captured in the Kanto area of Japan. J Vet Med Sci 1992;54: 461–463.
- Yoshida T, Sugimoto T, Sato Met al.Incidence of Listeria monocytogenes in wild animals in Japan. J Vet Med Sci 2000;62: 673–675.
- Hellstrom S, Kiviniemi K, Autio Tet al.Listeria monocytogenes is common in wild birds in Helsinki region and genotypes are frequently similar with those found along the food chain. J Appl Microbiol 2008;104: 883–888.
- Lyautey E, Hartmann A, Pagotto Fet al.Characteristics and frequency of detection of fecal Listeria monocytogenes shed by livestock, wildlife, and humans. Can J Microbiol 2007;53: 1158–1167.
- Poyart-Salmeron C, Carlier C, Trieu-Cuot Pet al.Transferable plasmid-mediated antibiotic resistance in Listeria monocytogenes. Lancet 1990;335: 1422–1426.
- Charpentier E, Courvalin P.Antibiotic resistance in Listeria spp. Antimicrob Agents Chemother 1999;43: 2103–2108.
- Li Q, Sherwood JS, Logue CM.Antimicrobial resistance of Listeria spp. recovered from processed bison. Lett Appl Microbiol 2007;44: 86–91.
- Tunkel AR, Hartman BJ, Kaplan SLet al.Practice guidelines for the management of bacterial meningitis. Clin Infect Dis 2004;39: 1267–1284.
- Gomez D, Azon E, Marco Net al.Antimicrobial resistance of Listeria monocytogenes and Listeria innocua from meat products and meat-processing environment. Food Microbiol 2014;42: 61–65.
- Wang X-M, Lü X-F, Yin Let al.Occurrence and antimicrobial susceptibility of Listeria monocytogenes isolates from retail raw foods. Food Control 2013;32: 153–158.
- Wang Y, Zhao A, Zhu Ret al.Genetic diversity and molecular typing of Listeria monocytogenes in China. BMC Microbiol 2012;12: 119.
- Wang Y, Jiao Y, Lan Ret al.Characterization of Listeria monocytogenes isolated from human Listeriosis cases in China. Emerg Microbes Infect 2015;4: e50.