Abstract
Coexistence of the hepatitis B surface antigen (HBsAg) and hepatitis B surface antibody (anti-HBs) is an uncommon phenomenon, and the underlying mechanisms remain largely unknown. Amino-acid (aa) substitution from glycine to arginine at aa 145 (G145R), in the major hydrophilic region, has been reported in patients with HBsAg and anti-HBs coexistence. However, there is limited knowledge about the clinical features and viral quasispecies characteristics associated with G145R mutant hepatitis B virus (HBV) infection. We herein describe the dynamic changes in the serological and virological markers in a case of hepatitis B with coexisting HBsAg and anti-HBs, caused by a G145R immune escape mutant (genotype C). Entecavir was administered during the 4th week after admission. Alanine aminotransferase peaked in the 16th week, while both the HBsAg and HBeAg declined rapidly. HBsAg clearance and hepatitis B e antigen (HBeAg)/hepatitis B e antibody (anti-HBe) seroconversion were achieved in the 36th week, and then entecavir was withdrawn. A follow-up of 96 weeks showed that HBV DNA remained undetectable and that anti-HBs was maintained above 100 mIU/mL. The quasispecies characteristics of the G145R mutant HBV were investigated via ultra-deep sequencing. The complexity and genetic distance of the S and RT regions were much higher in the 8th week than at baseline or in the 4th week. Moreover, the frequencies of mutations (L173P, Q181R and A184V) in cytotoxic T lymphocyte epitopes increased before entecavir treatment. These findings extend understanding of the evolution of HBV under host immune pressure and of the clinical outcomes of affected patients.
Emerging Microbes & Infections (2017) 6, e15; doi:10.1038/emi.2017.2; published online 22 March 2017
Introduction
The hepatitis B surface antigen (HBsAg) is a primary element used for the diagnosis of hepatitis B virus (HBV) infection, whereas hepatitis B surface antibody (anti-HBs) are often used as a serological marker of recovery. Nevertheless, the coexistence of HBsAg and anti-HBs has been reported,Citation1, Citation2, Citation3, Citation4 although the mechanisms underlying this uncommon phenomenon remain unclear. Mutations in the ‘α’ determinant of the major hydrophilic region (MHR), such as amino-acid (aa) substitution from glycine to arginine at aa 145 (G145R), have been reported in patients with coexistence of HBsAg and anti-HBs.Citation2, Citation5, Citation6, Citation7 However, there is limited knowledge about the clinical features and quasispecies characteristics associated with infection by G145R mutant HBV.
We herein describe the dynamic changes in the serological and virological markers in a case of hepatitis B with coexisting HBsAg and anti-HBs caused by a G145R immune escape mutant. Moreover, the quasispecies characteristics, including the frequency of mutations in full-length HBV genomes, were investigated using ultra-deep sequencing.
Materials and methods
Patient
The patient was a 90-year-old Chinese man who was admitted to the Geriatric Ward of Ruijin Hospital on 2 September 2014. He had not been vaccinated against HBV. A health checkup performed in October 2009 showed that he was negative for the HBsAg, hepatitis B e antigen (HBeAg) and hepatitis B e antibody (anti-HBe), whereas he was positive for the hepatitis B core antibody (anti-HBc, 1.41S/CO) and anti-HBs (129.57 mIU/mL, with a protective value above 10 mIU/mL). He had no history of known risk factors for HBV infection. He had been diagnosed with a gastric ulcer and diabetes three years before admission. The clinical and virological data from the patient were obtained during 2 years of follow-up. Informed consent was obtained from the patient.
Serological, biochemical and virological tests
Serological markers of HBV were quantified using an enzyme immunoassay kit (Murex Abbott, Chicago, IL, USA). HBV DNA was detected with a Cobas TaqMan HBV Test version 2.0 (lower limit of detection, 20 IU/mL; Roche Diagnostic, Basel, Switzerland). The alanine aminotransferase (ALT) level was determined according to the manufacturer’s protocol.
DNA extraction and direct PCR sequencing
Serum was obtained from the patient during the first (week 0; baseline), 4th and 8th weeks after admission. HBV DNA was extracted using a QIAamp DNA Blood Mini Kit (Qiagen, Tokyo, Japan). The S gene was amplified by nested PCR and sequenced after purification. The nucleotide sequences were aligned with reference sequences retrieved from GenBank, and the HBV genotype was identified through online software analysis (http://www.ncbi.nlm.nih.gov/projects/genotyping/formpage.cgi).
Ultra-deep sequencing
The entire HBV genomes were amplified with nine overlapping fragments (p1–p9), and then the concentration of the PCR products was measured using a Qubit dsDNA HS Assay Kit (Invitrogen, Carlsbad, CA, USA). A library of PCR products was established using a Nextera DNA Sample Prep Kit (Illumina, San Diego, CA, USA). Sequencing of the PCR products was performed using an Illumina (San Diego, CA, USA) Miseq system, according to the manufacturer’s paired-end 2 × 300 bp protocol. Image analysis and base calling were performed using Illumina CASAVA software version 1.8.2 with default parameters.
Treatment of sequencing data
Raw reads were pre-processed using CutAdaptCitation8 version 1.9.1 to cut adaptor sequences and trim low-quality reads (length <250 bp or base quality <30). Filtered read pairs were aligned to the HBV reference genome sequence (GenBank Accession Number: AB014381) with Bowtie2 version 2.2.6.Citation9 Samtools version 0.1.19 was used to generate coordinate-sorted bam files.Citation10 Sequencing errors induced by PCR and ultra-deep sequencing would complicate the analysis of mixed populations and result in inflated estimates of genetic diversity; therefore, we used ShoRAH 0.5.1 software,Citation11 which applied a probabilistic Bayesian approach to minimize the effects of errors.
Sequence characteristics and statistical analysis
The virus quasispecies heterogeneity was mainly evaluated on the basis of complexity and diversity. The quasispecies complexity was measured using normalized Shannon entropy (Sn). The Sn can be calculated with a previously described formula.Citation12, Citation13 The Sn of regions p1–p9 were calculated using Perl scripts in Perl 5.20. The quasispecies diversity was evaluated on the basis of the mean genetic distance (d). MEGACitation14 version 6.0 was used to calculate the genetic distance of all regions by using the Tamura three-parameter model.
Variations were detected and analyzed using Perl scripts. High-confidence variations with a frequency ≥1% of the total viral population were selected and analyzed. The quasispecies characteristics of the HBV genome were analyzed at baseline and during the 4th and 8th weeks as described previously.Citation15
Results
Dynamic changes in clinical characteristics during infection by G145R mutant HBV
At the time of admission, laboratory tests were positive for HBsAg (111.32 IU/mL), anti-HBs (771.71 mIU/mL), HBeAg (1179.194S/CO), anti-HBc IgM (1.21S/CO), and HBV DNA (5.82 log10 IU/mL) and indicated a slight elevation in ALT (43 IU/L; ). On 30 September 2014 (the 4th week after admission), tests revealed that the HBsAg, HBeAg and HBV DNA levels had increased significantly (895 IU/mL, 1475.496 S/CO and 6.84 log10 IU/mL, respectively), whereas the anti-HBs level had decreased to 502 mIU/mL. On the basis of the elevated HBV DNA and HBeAg levels, entecavir treatment was initiated at 0.5 mg per day with the patient’s consent.
Figure 1 The dynamic changes in serological markers, HBV DNA and ALT during infection by the HBV G145R immune escape mutant. Entecavir treatment was started during the 4th week. HBsAg became undetectable, and HBeAg/anti-HBe seroconversion was achieved during the 36th week, and then entecavir was withdrawn. aminotransferase, ALT; hepatitis B surface antigen, HBsAg; hepatitis B virus, HBV.
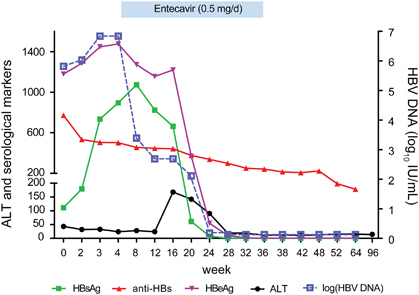
Interestingly, the ALT level increased to 168 IU/L during the 16th week, whereas the levels of HBsAg and HBeAg declined rapidly. HBV DNA became undetectable on 25 February 2015 (in the 24th week). At that time, the HBsAg and HBeAg levels had also decreased significantly (7.86 IU/mL and 54.728S/CO, respectively). HBsAg became undetectable, and HBeAg/anti-HBe seroconversion was achieved during the 36th week, and then entecavir was withdrawn. A follow-up of 96 weeks showed that HBV DNA remained undetectable and that the anti-HBs level was maintained above 100 mIU/mL. As shown in , the ALT level remained normal until the end of the 96-week follow-up.
Evolution of the viral quasispecies during G145R mutant HBV infection
A direct DNA sequence analysis was performed, and HBV genotype C with a G145R mutation was confirmed at the time of admission (). Accordingly, the patient was diagnosed with hepatitis B caused by a G145R immune escape mutant. Ultra-deep sequencing showed that the complexity and genetic distances within the S and RT regions in the 8th week were much higher than those at baseline and in the 4th week (, ).
Figure 2 The G145R mutation (A) and dynamic changes in major mutations in the S and RT regions (B). (A) Amino-acid substitution from glycine to arginine at aa 145 (G145R) in the major hydrophilic region of HBsAg was detected by direct sequencing at the time of admission. (B) The frequencies of major mutations at baseline and during the 4th and 8th weeks were detected using ultra-deep sequencing. Hepatitis B surface antigen, HBsAg.
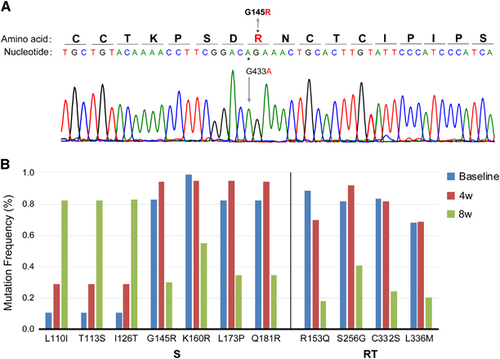
Figure 3 The complexity of the full-length HBV genome at baseline and during the 4th and 8th weeks. The complexity for each nucleotide was evaluated using ultra-deep sequencing. Entecavir treatment was initiated during the 4th week. Blue bars indicate the complexity for each nucleotide in the full-length HBV genome. Hepatitis B virus, HBV.
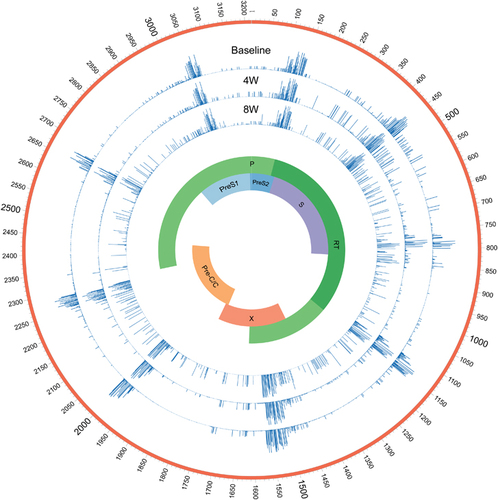
Table 1 The genetic distances at the nucleotide level at basline and during the 4th week and 8th week (10−3 substitutions/site)
Dynamic changes in the mutations in the HBV genome
The mutations with frequencies that changed during the course of treatment are shown in and the Supplementary Table S1. In addition to G145R, new dominant and minor mutations were detected. The frequencies of four mutations in the S region (G145R, K160R, L173P and Q181R), four mutations in the RT region (R153Q, S256G, C332S and L336M) and five mutations in the X region (S54P, L55F, R56P, L58P and C69P) decreased in the 8th week, whereas the frequencies of many mutations in the S, P and X regions clearly increased. The mutations with an increased frequency of detection were much more common than those with a decreased frequency (Supplementary Figure S1).
With regard to the ‘hot-spot’ mutations in the HBV genome, G1896A in the preC region and A1762T/G1764A in the basic core promoter region were studied. We also screened several mutations relevant to antiviral resistance, such as I169L, L180M, A181V, T184I, S202C, M204V/I, N236T and M250I in the RT region. None of these mutations were detected before or after the administration of entecavir.
Dynamic changes in the immune epitopes in the S region
As shown in Supplementary Figure S1, the frequencies of mutations (L173P, Q181R and A184V) in cytotoxic T lymphocyte (CTL) epitopesCitation16, Citation17 were increased before entecavir treatment, whereas mutations in the CTL domain aa 44–59 unexpectedly increased after entecavir treatment. Unlike mutations in the CTL domain, Y221C in the T-helper cell domainCitation16 exhibited a high frequency both before and after entecavir treatment.
Discussion
In the present study, the clinical and quasispecies characteristics associated with G145R immune escape mutant HBV infection were described in detail. The full-length HBV sequences obtained by ultra-deep sequencing showed that the complexity and genetic distances of the S and RT regions were much higher in the 8th week than at baseline and in the 4th week. Mutations in the MHR of HBsAg and T-cell epitopes may contribute to immune escape.
On the basis of the level of protective anti-HBs (129.57 mIU/mL) observed in 2009 and recent serological changes, hepatitis B caused by the G145R immune escape mutant was diagnosed. Infection with G145R mutant HBV may involve a clinical onset that is insidious, as in our subject, who had a slightly elevated ALT level. Of note, HBsAg and anti-HBs coexisted for 36 weeks in this patient. The prevalence of this atypical serological pattern in HBsAg-positive carriers is 2.9%–5%,Citation5, Citation18 and it tends to occur in older carriers (aged over 40 years) more often than in carriers who are HBsAg-positive but anti-HBs negative.Citation19 Another study has shown that patients with coexisting HBsAg and anti-HBs had higher proportions of HBV genotype C than patients with HBsAg-positive but anti-HBs-negative infections.Citation5 In addition, carriers with coexisting HBsAg and anti-HBs have high HBeAg- and HBV DNA-positive rates and are therefore infectious. A retrospective cohort study has shown that HBeAg and HBV DNA persist for several decades in most carriers, and their risk of hepatocellular carcinoma (HCC) is increased.Citation20 Fortunately, the HBsAg disappeared and HBeAg seroconversion was achieved in this patient, who eventually cleared the HBV and recovered.
The mechanisms underlying the simultaneous appearance of HBsAg and anti-HBs remain controversial. The presence of amino-acid substitutions in the MHR of HBsAg, such as G145R and I126T, can change the immunogenicity of HBsAg, thus resulting in HBsAg/anti-HBs coexistence and immune escape. Consequently, these mutations can also lead to the diagnostic failure of commercial assays for HBsAg, as well as to the prophylactic failure of immunoglobulin or vaccines. The G145R mutation in the S region is likely to be responsible for weak recognition by anti-HBs.Citation21, Citation22 Because the G145R mutation alters the projecting loop of the ‘α’ determinant, pre-existing neutralizing antibodies cannot adequately recognize the changed epitope. Other studies have suggested that exposure to different subtypes of HBV accounts for the anomalous serological changes observed in these patients.Citation3, Citation19, Citation23 Zhang et al.Citation3 have reported that the subtype-specific anti-HBs in these patients are directed to HBsAg subtypes other than the coexisting subtype and are unable to neutralize the coexisting HBsAg. Owing to a high replication rate and lack of proofreading activity during reverse transcription, HBV exists as a spectrum of variants called quasispecies.Citation12, Citation13 These variants are genetically distinct but closely related. Because of their different adaptability, the presence and type of quasispecies are related to the outcome of HBV infection. In the present study, data from ultra-deep sequencing revealed that mutants harboring different mutations, including G145R and I126T, coexisted in the patient. Furthermore, the ratio of coexisting mutants varied during the infection. These findings are consistent with the two aforementioned hypotheses and suggest that mutations in the S gene, as well as different subtypes of HBV, may play important roles in the coexistence of HBsAg and anti-HBs.
Increasing evidence indicates that the characteristics of HBV quasispecies, including their complexity and genetic distances, are related to the outcome of HBV infection and antiviral therapy.Citation12, Citation13, Citation24, Citation25, Citation26 However, the characteristics of the entire genomes of HBV quasispecies determined using ultra-deep sequencing during G145R mutant infection have not yet been reported. The quasispecies heterogeneity within the S and RT regions had not changed significantly during the 4th week compared with baseline but clearly increased 1 month after the onset of administration of entecavir.
Importantly, ultra-deep sequencing provided insights into the selection process of immune escape mutants during HBV infection and entecavir treatment. Accompanying high levels of HBV DNA, the frequencies of G145R and other mutations (K160R, L173P, Q181R in the S region and R153Q, S256G, C332S, L336M in the RT region) were high both at baseline and in the 4th week. Interestingly, under antiviral therapy, both the HBV DNA level and the frequencies of the aforementioned mutations declined during the 8th week, as expected. In contrast, the frequencies of L110I, T113S and I126T, which may be compensatory mutations, increased in the S region. Compared with other antiviral agents, entecavir is more potent, and it causes higher selective pressure on HBV quasispecies, thus possibly accounting for the elevated quasispecies complexity observed in this study. High quasispecies complexity, which is associated with a large reservoir of variants and more complementary interactions among variants, means that there is high quasispecies adaptability.Citation12 In addition, the interplay between different mutants may facilitate the replication of the viral population.Citation27 Thus, quasispecies with higher complexity should theoretically tend to breach genetic barriers, thus leading to drug resistance. Nevertheless, entecavir has a higher genetic barrier to drug resistance, and the use of entecavir for therapy can prevent the emergence of many of the known mutations leading to drug resistance. To date, the consequences of this selection and evolution remain largely unknown.
A previous study has indicated that mutations in the S region mainly occur in the N-terminal region (aa 1–99) and the MHR (aa 100–169), which contain the ‘α’ determinant (aa 124–147), rather than the C-terminal region (aa 170–226).Citation5 Consistently with results from a previous study,Citation5 the data from ultra-deep sequencing in the present study showed that mutations in the S region were mainly located in the MHR and N-terminal region, both before and after antiviral therapy. The most frequently observed mutations in the S region were located at aa 126, 129, 130, 133, 145 and 181.Citation2, Citation6, Citation22 The G145R mutation in the MHR, which is one of the most prevalent immune escape mutations, can impair HBsAg secretionCitation28 and has been reported in patients with advanced liver diseases and even liver cancer.Citation15 In addition, the Q181R mutation has been recently reported in a new vaccine escape mutant.Citation22 Some of these mutations have been reported in cases of occult HBV infection.Citation2, Citation5, Citation6
In summary, our study shows the dynamic changes in the clinical features and quasispecies characteristics during infection with G145R mutant HBV. In addition to mutations in the MHR of HBsAg, mutations in T-cell epitopes may contribute to immune escape. These findings obtained by ultra-deep sequencing extend understanding of the evolutionary pattern of HBV under host immune pressure and of the clinical outcomes of patients.
Supplementary Figure S1
Download PDF (5.7 MB)Supplementary Table S1
Download MS Excel (21 KB)Acknowledgments
This work was supported by grants from the Major National Projects for Infectious Diseases (2012ZX10002007, 2013ZX10002001), the National Natural Science Foundation of China (81371860), the State Major Basic Research Program (973) of China (2012CB519000) and the Special Research Fund of the Ministry of Health for the Non-Profit Sector (201302010).
Supplementary Information for this article can be found on the Emerging Microbes & Infections website (http://www.nature.com/emi)
References
- Galati G, De Vincentis A, Vespasiani-Gentilucci Uet al.Coexistence of HBsAg and HBsAb in a difficult-to-treat chronic hepatitis B: loss of HBsAg with entecavir plus tenofovir combination. BMC Gastroenterol 2014;14: 94.
- Lada O, Benhamou Y, Poynard Tet al.Coexistence of hepatitis B surface antigen (HBs Ag) and anti-HBs antibodies in chronic hepatitis B virus carriers: influence of 'a' determinant variants. J Virol 2006;80: 2968–2975.
- Zhang JM, Xu Y, Wang XYet al.Coexistence of hepatitis B surface antigen (HBsAg) and heterologous subtype-specific antibodies to HBsAg among patients with chronic hepatitis B virus infection. Clin Infect Dis 2007;44: 1161–1169.
- Yu DM, Li XH, Mom Vet al.N-glycosylation mutations within hepatitis B virus surface major hydrophilic region contribute mostly to immune escape. J Hepatol 2014;60: 515–522.
- Liu W, Hu T, Wang Xet al.Coexistence of hepatitis B surface antigen and anti-HBs in Chinese chronic hepatitis B virus patients relating to genotype C and mutations in the S and P gene reverse transcriptase region. Arch Virol 2012;157: 627–634.
- Bian T, Yan H, Shen Let al.Change in hepatitis B virus large surface antigen variant prevalence 13 years after implementation of a universal vaccination program in China. J Virol 2013;87: 12196–12206.
- Thakur V, Kazim SN, Guptan RCet al.Transmission of G145R mutant of HBV to an unrelated contact. J Med Virol 2005;76: 40–46.
- Turner FS.Assessment of insert sizes and adapter content in fastq data from NexteraXT libraries. Front Genet 2014;5: 5.
- Langmead B, Salzberg SL.Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9: 357–359.
- Li H, Handsaker B, Wysoker Aet al.The Sequence alignment/Map format and SAMtools. Bioinformatics 2009;25: 2078–2079.
- Zagordi O, Klein R, Daumer Met al.Error correction of next-generation sequencing data and reliable estimation of HIV quasispecies. Nucleic Acids Res 2010;38: 7400–7409.
- Liu F, Chen L, Yu DMet al.Evolutionary patterns of hepatitis B virus quasispecies under different selective pressures: correlation with antiviral efficacy. Gut 2011;60: 1269–1277.
- Chen L, Zhang Q, Yu DMet al.Early changes of hepatitis B virus quasispecies during lamivudine treatment and the correlation with antiviral efficacy. J Hepatol 2009;50: 895–905.
- Tamura K, Stecher G, Peterson Det al.MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 2013;30: 2725–2729.
- Yamani LN, Yano Y, Utsumi Tet al.Ultradeep sequencing for detection of quasispecies variants in the major hydrophilic region of hepatitis B virus in Indonesian patients. J Clin Microbiol 2015;53: 3165–3175.
- Khedive A, Norouzi M, Ramezani Fet al.Hepatitis B virus surface protein mutations clustered mainly in CTL immune epitopes in chronic carriers: results of an Iranian nationwide study. J Viral Hepat 2013;20: 494–501.
- Honorati MC, Dolzani P, Mariani Eet al.Epitope specificity of Th0/Th2 CD4+ T-lymphocyte clones induced by vaccination with rHBsAg vaccine. Gastroenterology 1997;112: 2017–2027.
- Pancher M, Desire N, Ngo Yet al.Coexistence of circulating HBsAg and anti-HBs antibodies in chronic hepatitis B carriers is not a simple analytical artifact and does not influence HBsAg quantification. J Clin Virol 2015;62: 32–37.
- Pu Z, Li D, Wang Aet al.Epidemiological characteristics of the carriers with coexistence of HBsAg and anti-HBs based on a community cohort study. J Viral Hepat 2016;23: 286–293.
- Seo SI, Choi HS, Choi BYet al.Coexistence of hepatitis B surface antigen and antibody to hepatitis B surface may increase the risk of hepatocellular carcinoma in chronic hepatitis B virus infection: a retrospective cohort study. J Med Virol 2014;86: 124–130.
- Wu C, Deng W, Deng Let al.Amino acid substitutions at positions 122 and 145 of hepatitis B virus surface antigen (HBsAg) determine the antigenicity and immunogenicity of HBsAg and influence in vivo HBsAg clearance. J Virol 2012;86: 4658–4669.
- Ye Q, Shang SQ, Li W.A new vaccine escape mutant of hepatitis B virus causes occult infection. Hum Vaccin Immunother 2015;11: 407–410.
- Ding F, Yu HG, Li YXet al.Sequence analysis of the HBV S protein in Chinese patients with coexisting HBsAg and anti-HBs antibodies. J Med Virol 2015;87: 2067–2073.
- Han Y, Gong L, Sheng Jet al.Prediction of virological response by pretreatment hepatitis B virus reverse transcriptase quasispecies heterogeneity: the advantage of using next-generation sequencing. Clin Microbiol Infect 2015;21: e791–e798.
- Lavocat F, Deny P, Pichoud Cet al.Similar evolution of hepatitis B virus quasispecies in patients with incomplete adefovir response receiving tenofovir/emtricitabine combination or tenofovir monotherapy. J Hepatol 2013;59: 684–695.
- Zhou B, Dong H, He Yet al.Composition and interactions of hepatitis B virus quasispecies defined the virological response during telbivudine therapy. Sci Rep 2015;5: 17123.
- Vignuzzi M, Stone JK, Arnold JJet al.Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature 2006;439: 344–348.
- Amini-Bavil-Olyaee S, Vucur M, Luedde Tet al.Differential impact of immune escape mutations G145R and P120T on the replication of lamivudine-resistant hepatitis B virus e antigen-positive and -negative strains. J Virol 2010;84: 1026–1033.
