Abstract
A large-scale survey was conducted in domestic animal populations from 2011 to 2015 in Qingyang, China. A total of 448,398 animals from different districts of Qingyang were tested for the presence of Brucella-specific antibodies using the Rose Bengal Plate Test (RBPT) and the Standard Agglutination Test (SAT). From 2011 to 2015, the yearly average positive rates were between 0.04 and 4.75% in the eight counties tested. In addition, the prevalence rates were between 0 and 9.96% in these eight counties. Sheep was the dominant host of Brucella in Qingyang, and the prevalence rate in sheep (2.74%) was higher than those in the other animals tested. Identification of 10 Brucella isolates from sheep confirmed that the epidemic strains were B. melitensis biovar 3 (n = 9) and B. melitensis biovar 1 (n = 1). MLVA-11 (multiple-locus variable-number tandem repeat analysis) analysis of the 10 isolates showed three genotypes: genotype 116 (n = 8), genotype 115 (n = 1) and genotype 136 (n = 1). Furthermore, analysis of the whole-genome sequences of the representative B. melitensis strain QY1 indicated that this isolate was closely related to isolates from China and India. The results of serum epidemiology confirmed that the region of northern Qingyang was a critical Brucella epidemic area and that the disease showed a rising trend, especially from 2013 to 2015. An analysis of the isolate genotypes suggested that sheep brucellosis mainly resulted from conventional B. melitensis (East Mediterranean group), although the external strain (American group) also occurred in Qingyang.
Introduction
Brucellosis, caused by Brucella spp., is a zoonotic bacterial disease that has serious implications for human and animal healthCitation1. It is a highly contagious disease that affects livestock such as cattle, sheep and pigs. The symptoms include abortion, infertility, decreased production and lameness in animals and undulating fever with arthralgia in humansCitation2. Recently, this disease has re-emerged in some regions of ChinaCitation3–Citation6. From an epidemiological perspective, the major cases in animals have been sheep infected by B. melitensis. Human brucellosis has re-occurred in the past several yearsCitation7,Citation8. In Qingyang, four cases were detected from 2006 to 2008, while from 2009 to 2012, the number of positively detected Brucella cases was between 25 and 57. In 2013 and 2014, however, the cases of human brucellosis numbered 161 and 457, respectivelyCitation9. Among animal cases, from 2009 to 2013, the seroprevalence was <1%, although brucellosis was already occurring in domestic animalsCitation10.
Over the past decade, the number of breeding stock of both cattle and swine were approximately 400,000 in Qingyang. However, sheep farming was developed vigorously with main farmers feed. From 2007 to 2013, more than two million sheep were bred, and the number of sheep breeding stock was over 4 million in both 2014 and 2015 in Qingyang. From the beginning of this century to 2016, although animal brucellosis occurred in an increasing number of cases, the local control strategy was to cull the infected animals that presented as serologically Brucella positive. After 2016, vaccination was required in the area because of changes in government control policy, which were based on an assessment of the development of brucellosis in animals. Therefore, detailed knowledge of the epidemiological situation has become very important for the vaccination program and for assessment of effective prevention. This study was aimed at understanding the enzootic situation of Brucella in livestock and the characteristics of epidemics. Comprehensive Brucella seroepidemiological surveys were performed in livestock epidemics of brucellosis from 2011 to 2015 in Qingyang, China. To further illuminate the molecular features of epidemic Brucella, bacterial isolation and whole-genome sequencing were also performed.
Results
Domestic brucellosis in Qingyang from 2011 to 2015
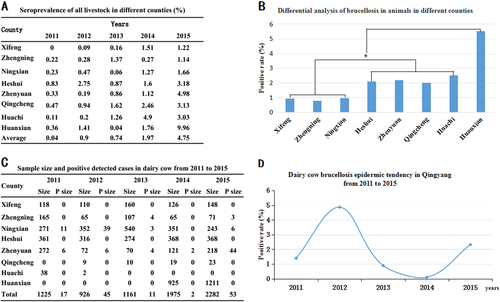
Of a total of 448,398 serum samples from different animals, 11,654 (2.6%) were anti-Brucella positive. The yearly positive rates in swine, yellow cattle (native breed), dairy cows (Holstein), sheep and dogs were 0%, 0.04%, 1.66%, 2.74% and 0.14%, respectively. Sheep and dairy cows with Brucella had higher infection trends, although the positive rates in dairy cows dipped to a downward trend between 2013 and 2014. Compared with that in sheep and dairy cows, brucellosis in yellow cattle and dogs maintained sporadic onset, and no cases were found in swine (Table ). From 2011 to 2015, the average positive rates in each year were from 0.04 to 4.75% in eight counties. In addition, the yearly prevalence rates were among 0 to 9.96% in these eight counties, and the disease emerged with an obvious rising trend, especially in the years 2013 and 2015. However, brucellosis in animals showed regional differences (Figs. ). Overall, domestic brucellosis in Qingyang predominantly infected sheep. Huanxian, located in the north Qingyang district, was a serious animal Brucella epidemic area. In the years 2013 and 2014, 6 (0.04%) of the 13,793 total yellow cattle were brucellosis positive, and 0.14% (1/698) seroprevalence was detected in dogs (Table ).
Sample size and positive number of different animals from 2011 to 2015 years
a The positive rates of the different counties in each year. b Differential analysis of brucellosis in different counties from 2011 to 2015. Bar, the average seroprevalence from 2011 to 2015; *Significant difference at P < 0.05. c Sample sizes and positively detected cases in the dairy cow brucellosis epidemic in Qingyang from 2011 to 2015. d Dairy cow brucellosis epidemic tendency in Qingyang from 2011 to 2015
Dairy cow brucellosis in Qingyang
Serological examination revealed that dairy cow brucellosis mainly emerged in the southern region of Qingyang, including Ningxian, Zhenyuan and Zhengning counties, whereas no dairy cow Brucella cases were found in the northern region of Qingyang. The yearly prevalence rates of Brucella-infected dairy cows was from 0.89 to 4.86% from 2011 to 2015 without incidence trends, and the highest positive detection rate was 20.12% in Zhenyuan in 2015 (Figs. ).
Sheep brucellosis in Qingyang
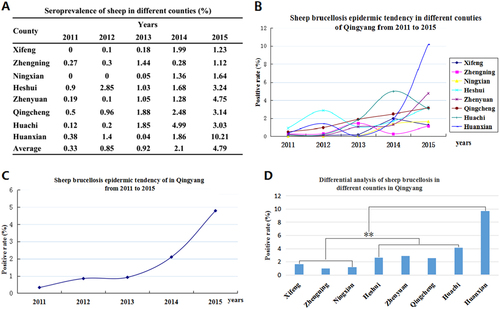
To clarify the situation of Brucella in sheep in Qingyang district, an analysis of the epidemic in eight counties showed that the Brucella prevalence in each year ranged from 0 to 10.21% (Fig. ), and the average was found to be 0.33 to 4.79% from 2011 to 2015 (Figs. ). Furthermore, the prevalence of Brucella showed an obvious rising trend, and the disease displayed a significant regional difference (Figs. ). According to the geographical locations of the eight counties, Brucella in sheep showed an increasing gradient from south to north. Three southern counties, Zhengning, Ningxian and Xifeng, had lower positive rates (<2%) in sheep, but the positive rate in northern Huanxian was the highest (9.7%). In the other counties, including Zhengyuan, Qingcheng, Huachi and Heshui, the positive rates of brucellosis were from 2.65 to 4.17% (Fig. ). Although the average seroprevalence in Huanxian was significantly higher than those in other counties, no regional difference (P > 0.05) was displayed because of the outbreak in 2015.
a The positive rates of the different counties in each year from 2011 to 2015. b Sheep brucellosis epidemic tendency in Qingyang from 2011 to 2015. c Sheep brucellosis epidemic tendencies in different counties in Qingyang from 2011 to 2015. d Differential analysis of the seroprevalence in different counties in Qingyang. Bar, the average seroprevalence from 2011 to 2015; **Very significant difference at P < 0.01
Isolation and identification of Brucella strains in sheep
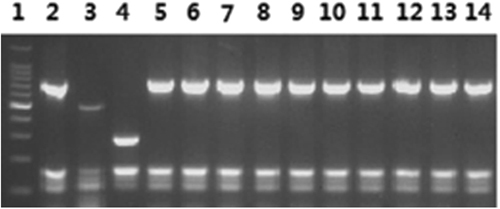
From 55 spleens of aborted samples, a total of 10 isolates were obtained. Specifically, five and two isolates were isolated from Huanxian and Zhenyuan, and one isolate was isolated from each of Huachi, Heshui and Zhengning. Biochemical testing and AMOS-PCR showed that nine biovars were identified, of which one was B. melitensis biovar 1 and nine were B. melitensis biovar 3 (n = 1; Fig. ).
Molecular epidemiological tracking of isolates from sheep
Analysis of MLVA-16 (multiple-locus variable-number tandem repeat analysis) loci revealed diverse genotypes among the 10 isolates in Qingyang. B. melitensis biovar 3 was the predominant biovar in the examined areas. According to the panel 1 markers, all isolates were clustered into four known genotypes: 45 (1-5-3-12-2-2-3-2; n = 1), 63 (1-5-13-2-3-3-2; n = 3), 42 (1-5-3-13-2-2-3-2; n = 5) and 47 (3-4-2-13-4-2-3-3; n = 1). The MLVA-11 assay involved 11 loci including Bruce06, Bruce08, Bruce11, Bruce12, Bruce18, Bruce19, Bruce21, Bruce42, Bruce43, Bruce45 and Bruce55. The results of the MLVA-11 analysis showed that the 10 isolates included three genotypes: genotype 116 (n = 8), genotype 115 (n = 1) and genotype 136 (n = 1). To track the source of the infection strains and their evolutionary relationships, MLVA-11 was used to analyze and compare the isolates in Qingyang with B. melitensis strains in other provinces, including Shandong, Inner Mongolia, Xinjiang, Ningxia, Hebei, Heilongjiang, Henan, Qinghai, Chongqing, Shanxi, Sichuan and Zhejiang provincesCitation8. Comparing these 10 isolates with different B. melitensis strains from sheep based on MLVA-11, it was shown that the isolates located in three relatively independent clusters (clusters A, B and C) represented different B. melitensis strains from another region (Fig. and Table S1).
Phylogenetic and evolutionary relationships of B. melitensis QY1
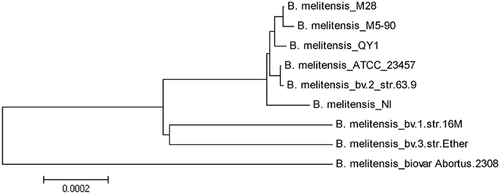
Analysis of the whole-genome sequences showed that B. melitensis QY1 has two circular chromosomes with 2,125,648 bp in chromosome I and 1,185,604 bp in chromosome II. The results of a phylogenetic tree based on the whole genome demonstrated that B. melitensis QY1 is close to B. melitensis M28, B. melitensis NI and B. melitensis M5-90, which originated in China, as well as B. melitensis bv. 3 str. Ether, a strain that was isolated from India (Fig. ).
The evolutionary history was inferred using the neighbor-joining method. The optimal tree (with branch length sum = 0.00316668) is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree
Discussion
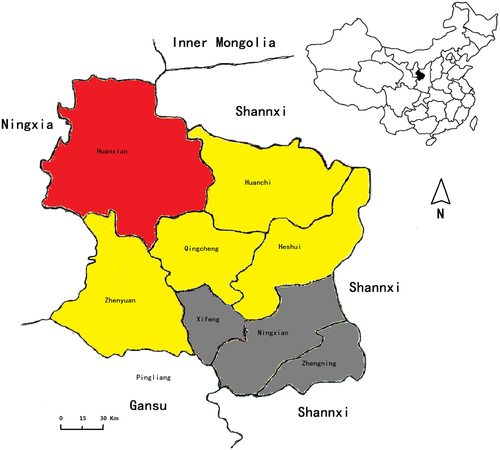
Qingyang is located at the intersection of Gansu, Shaanxi and Ningxia provinces and is near Inner Mongolia. The latter two provinces have very large livestock populations with high prevalences of BrucellaCitation3,Citation11. In the present study, we used serological methods to investigate animal brucellosis from 2011 to 2015 in Qingyang, China. The results of seroprevalence assays confirmed that Brucella spp. was mainly epidemic in sheep and dairy cows, whereas other animals had a low incidence. The results also showed that the disease has attained an obvious rising trend. Similarly, the number of human cases of brucellosis has also increasedCitation9,Citation12,Citation13. These results imply that the prevalence of brucellosis in animals, especially in sheep, has caused an increase in the human disease. In contrast, although Brucella infected a large number of sheep and dairy cows, yellow cattle presented very few cases.
From 2011 to 2015, brucellosis in dairy cows emerged without incidence trends in Qingyang. There are two possible explanations for this phenomenon. The breeding stock of dairy cows was <3000, so the disease was easy to control. In addition, the local control strategy was to cull infected animals, which largely prevented the expansion of Brucella in the short term. From an epidemiological perspective, sheep brucellosis emerged broadly in Qingyang, and dairy cow brucellosis occurred as outbreaks in some dairy farms. It was implied that brucellosis in dairy cows came from infected sheep.
Currently, the number of sheep breeding stock is over 4 million in Qingyang, and almost half of the sheep were raised in Huanxian. From 2011 to 2014, the seroprevalence of brucellosis in Huanxian was <2%, but an outbreak of the disease emerged in the year 2015. The most important factors may be convenient transportation and the prevalence of Brucella in most regions of China, as well as monitoring of animals with Brucella. Meanwhile, in the past few years, the prevalence of brucellosis in animals and humans has increased each year in Ningxia province, which is adjacent to northern HuanxianCitation11. This result may be explained by the fact that frequent livestock exchange occurs between these areas. The outbreak of brucellosis in 2015 further reflected a lack of control in the movement of infected animals between the regions and the supervision of the market. This hypothesis suggests that inspection measures to ensure the quarantine of livestock exported from endemic regions should be strengthenedCitation7.
To clarify the molecular features of sheep Brucella in Qingyang, 10 isolates were revived from Huanxian, Zhenyuan, Huachi, Heshui and Zhengning. Genotyping by MLVA-11 showed that the isolates were divided into three relatively independent clusters (cluster A, B and C; Fig. ). Cluster A, containing five isolates, was very close to major strains that were responsible for the dominant form of sheep brucellosis in China. However, the isolates in Cluster B were closely related to a few strains isolated from Ningxia, Inner Mongolia, Heilongjiang and ShandongCitation8. An isolate in Cluster C belongs to B. melitensis biovar 1, which was much closer to Brucella strains isolated from Hainan, China, in 2013Citation8. The results of MLVA-11 confirmed that major isolates were genotype 116 (n = 8), a common genotype derived from Brucella strains in the East Mediterranean. Moreover, this genotype was also responsible for the vast majority of Brucella infections in China. However, an isolate (genotype 136) from an American lineage was identified and observed in this study. Based on the analysis of these results, it is suggested that sheep brucellosis in Qingyang occurred not only due to a primary epidemic focus but also because of invasion by external bacterial strains with the introduction of sheep.
B. melitensis QY1, as a representative strain, was sequenced and analyzed. The whole genome was 3,311,252 bp in size. This study compared B. melitensis QY1 with B. melitensis M28, B. melitensis M5-90, B. melitensis NI, B. melitensis bv. 1 str. 16M, B. melitensis bv. 3 str. Ether, B. melitensis biovar abortus 2308 and B. melitensis ATCC 23457. The results of comparative genomics confirmed that B. melitensis QY1 was a dominant pathogenic strain in sheep and had a high identity to Brucella strains from China.
Sheep brucellosis re-emerged in Qingyang with rising trends. Therefore, a wide range of vaccination strategies were adopted starting in 2016. China has applied a vaccination program using live attenuated strains S2 (B. suis biovar 1 S2) and M5 (B. melitensis M5-90)Citation14–Citation16, but the problem is to distinguish positive sera due to vaccination from positive sera due to field strain infection. Modified live attenuated vaccines have been developed using differential diagnosis, but so far there are no commercial applications. Studies of molecular epidemiology and strain characteristics can provide support for vaccine selection and development.
Materials and methods
Ethics statement
This study involves the investigation and isolation of Brucella spp. using serology and modern typing methods. All animal experiments in the study were approved by the Animal Ethics Committee of Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, and were conducted strictly according to the guidelines.
Collection of samples and testing
Serum samples were collected from livestock in Qingyang district, China, from March through July in 2011 to 2015. Samples of aborted sheep fetuses were collected from April through July in both 2015 and 2016. From 2011 to 2015, a total of 448,398 serum samples, including swine (4237), yellow cattle (16,433), dairy cow (7569), sheep (419,461) and dog (698), were collected from Qingyang district, Gansu province, China (Table ). These samples were collected from eight counties, including Xifeng (32,478), Zhengning (30,904), Ningxian (56,228), Heshui (62,906), Zhenyuan (44,576), Qingcheng (48,212), Huachi (75,396) and Huanxian (98,213) (Fig. , Table and Table S2). The sample collection rates in sheep were from 2 to 5% (proportion of total population), the dairy cows were fully sampled, and sampling rates in the other livestock were higher than 20%. All sample collection was performed to cover the full area of Qingyang district, which was sampled randomly. All serum samples were transported to the laboratory, where they were stored at −20 °C until processing and detection by Rose Bengal Plate Test (RBPT), and the positive serum samples were reconfirmed by a Standard Agglutination Test (SAT)Citation17 (all the reagents were purchased from China Institute of Veterinary Drug Control).
Isolation of Brucella
To study the molecular epidemiological characters, 55 spleens of aborted sheep were collected at random from eight counties and subjected to a brucellosis purification progress. Spleens were crushed and cultured on Brucella serum dextrose agar composed of Brucella medium base (supplemented with Brucella selective antibiotic, OXOID, England), and 5–10% heat-inactivated horse serum. Plates were incubated with and without 5–10% carbon dioxide at 37 °C after inoculation with sample materials. The plates were examined after 3–15 days for bacterial growth. A single clone was chosen for identification.
Identification of isolates
The obtained single bacterial clones were identified by biochemical testing according to the standard strain identification methodCitation17–Citation19. The carbon dioxide (CO2) requirement was tested on Brucella serum dextrose agar with and without CO2 during the first isolation. Agglutination by A, M and R monospecific antisera were detected by mixing the antisera with the isolate after dilution of the colony (all reagents were purchased from The Chinese Center for Disease Control and Prevention). Total genomic DNA was extracted using a DNeasy Blood & Tissue Kit (Qiagen, Germany) according to the manufacturer’s instructions. The DNA extracted from all isolates was stored at −20 °C. AMOS-PCR was performed as previously describedCitation20,Citation21.
Molecular epidemiological tracking of isolates by MLVA-16
MLVA was performed as described previouslyCitation22–Citation24. Sixteen primer pairs were included in panel 1 (bruce06, bruce08, bruce11, bruce12, bruce42, bruce43, bruce45 and bruce55), panel 2A (bruce18, bruce19 and bruce21), and panel 2B (bruce04, bruce07, bruce09, bruce16 and bruce30). Polymerase chain reaction (PCR) amplification was performed in a 50 μL reaction system, containing 1 μL DNA template, 1 μL of each primer, 25 μL of 2 × Premix Taq, and 22 μL of ddH2O. The reaction conditions were as follows: initial denaturation at 94 °C for 5 min; followed by 40 cycles of 94 °C for 30 s, 60 °C for 30 s and 72 °C for 1 min. At last, the samples were incubated for an additional 10 min at 72 °C and were then stored at 4 °C. Five microliters of each PCR product from panel 1 was loaded onto a 2% agarose gel with 0.5% ethidium bromide, whereas those from panels 2A and 2B were loaded into 3% agarose gels also containing 0.5% ethidium bromide. The bands were visualized under UV light and photographed. The PCR products for the 16 loci were denatured and resolved by capillary electrophoresis on an ABI Prism 3130 automated fluorescent capillary DNA sequencer (Applied Biosystems). Fragments were sized following comparison with a ROX (carboxy-X-rhodamine)-labeled molecular ladder (MapMarker 1000; BioVentures Inc., Murfreesboro, TN, USA) and Gene Mapper software version 4.0 (Applied Biosystems). Band intensities were estimated using BioNumerics version 7.6 (Applied Maths, Belgium), and repeat units were then obtained according to the published allele numbering systemCitation22,Citation23. Clustering analysis was performed using the same software based on the categorical coefficient and unweighted pair group using arithmetic averages method. The resulting genotypes were compared using the web-based Brucella 2016 MLVA database (http://mlva.u-psud.fr/) and published papersCitation23. Advanced clustering analysis was based on the MLVA for categorical data in the BioNumerics system.
Genomic sequencing and evolutionary relationships of B. melitensis QY1
A dominant epidemic isolate, B. melitensis QY1, was used for whole-genome sequencing. Genomic DNA was extracted with the sodium-dodecyl sulphate (SDS) method. The harvested DNA was detected by agarose gel electrophoresis and quantified by Qubit (Life Qubit ® Fluorescence Quantitative Instrument, USA). The genome of B. melitensis QY1 was sequenced by single-molecule real-time (SMRT) technology. Sequencing was performed at Beijing Novogene Bioinformatics Technology Co., Ltd. The low-quality reads were filtered by SMRT 2.3.0Citation25,Citation26, and the filtered reads were assembled to generate one contig without gaps (accession nos. CP022204.1-CP022205.1)Citation27. The evolutionary relationships were evaluated in comparison with B. melitensis M28, B. melitensis M5-90, B. melitensis NI, B. melitensis bv. 1 str. 16M, B. melitensis bv. 3 str. Ether, B. melitensis biovar abortus 2308 and B. melitensis ATCC 23457Citation28 (Table S3). Based on the whole genomes of nine selected B. melitensis strains, the amino-acid sequences of core genes were analyzed by the CD-HIT software for rapid clustering of similar proteins with a threshold of 50% pairwise identity and 0.7 length difference cutoff. A phylogenetic tree was constructed from the orthologous gene sequences by TreeBeST using the neighbor-joining method, and the number of bootstrap samples was 1000Citation29,Citation30.
Statistical analysis
The data were transferred to Microsoft Excel and evaluated using a two-tailed unpaired t-test to compare the two groups; P-values <0.05 were considered to be significantly different.
supplement Table S1(DOC 399 kb)
Download MS Word (399 KB)supplement Table S2(DOC 53 kb)
Download MS Word (53 KB)supplement Table S3(DOC 30 kb)
Download MS Word (30 KB)Acknowledgements
This study was supported in part by the National Key Research and Development Plan (2016YFD0500907), Fundamental Research Funds for CAAS (1610312017014), Gansu Province Scientific and Technical Supporting Program (no. 1304NKCA162) and Gansu Province Agricultural Biotechnology Research and Application Development Project (no. GNSW-2013-27).
Authors' contributions
X.C., S.L., Z. Li, Z. Liu, J.M., Z. Lou, J.Z. and Y.L. performed the experiments; X.C. and Z. Li analyzed the data; X.C., S.L., Z.J. and B.F. initiated and designed the research; X.C. wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41426-018-0060-y).
References
- Hasanjani RoushanMREbrahimpourSHuman brucellosis: an overviewCasp. J. Intern. Med.20156 46 47
- CastanoMJSoleraJChronic brucellosis and persistence of Brucella melitensis DNAJ. Clin. Microbiol.2009472084208910.1128/JCM.02159-082708509
- HouQModeling the transmission dynamics of sheep brucellosis in Inner Mongolia autonomous region, ChinaMath. Biosci.2013242515810.1016/j.mbs.2012.11.012
- ZhangJPrediction and control of brucellosis transmission of dairy cattle in Zhejiang Province, ChinaPLoS. ONE.20149e10859210.1371/journal.pone.01085924227660
- MorenoERetrospective and prospective perspectives on zoonotic brucellosisFront. Microbiol.2014521310.3389/fmicb.2014.002134026726
- ChenSIncreasing threat of brucellosis to low-risk persons in urban settings, ChinaEmerg. Infect. Dis.20142012613010.3201/eid2001.1303243884711
- LiuZGMLVA genotyping characteristics of human Brucella melitensis isolated from Ulanqab of Inner Mongolia, ChinaFront. Microbiol.2017865241320
- TianGZMulti-locus variable-number tandem repeat analysis of Chinese Brucella strains isolated from 1953 to 2013Infect. Dis. Poverty2017610.1186/s40249-017-0296-05412030
- Liu, S. L. & Hui, Y. L. Epidemiological analysis of Brucellosis in Qingyang of Gansu Province from 2006 to 2014 (Chinese). Bull. Dis. Control Prev. 30 37–39 (2015).
- Lu, Y. W., Huo, F. Y. & Li, S. E. Epidemiological Investigation of Brucellosis in Qingyang from 2009 to 2013 (Chinese). J. Animal Sci. Veterinary Med.33 73–76 (2014).
- Guo, Y. H. & Zhang, T. Supervision and analysis of Brucellosis in Yanchi of Ningxia province from 2010 to 2014 (Chinese). J. Ningxia Med. Univ.37 1078–1080 (2015).
- JiaBBrucella endocarditis: clinical features and treatment outcomes of 10 cases from Xinjiang, ChinaJ. Infect.20177451251410.1016/j.jinf.2017.01.011
- ChenYChanges of predominant species/biovars and sequence types of Brucella isolates, Inner Mongolia, ChinaBMC. Infect. Dis.20131310.1186/1471-2334-13-5143819263
- ShangD[Progress in the study of prevention and control of Brucellosis in China in last 50 years]Zhonghua. Liu. Xing. Bing. Xue. Za. Zhi.2000215557
- XinXOrally administrable brucellosis vaccine: Brucella suis strain 2 vaccineVaccine1986421221610.1016/0264-410X(86)90131-3
- VergerJMGrayonMZundelELechopierPOlivier-BernardinVComparison of the efficacy of Brucella suis strain 2 and Brucella melitensis Rev. 1 live vaccines against a Brucella melitensis experimental infection in pregnant ewesVaccine19951319119610.1016/0264-410X(95)93135-V
- Bovine, B. The World Assembly of Delegates of the OIE. Ch. 11. 3. Institut National de la Recherche Afronomique. Paris, France, (2009).
- Al DahoukSTomasoHNocklerKNeubauerHFrangoulidisDLaboratory-based diagnosis of brucellosis--a review of the literature. Part I: techniques for direct detection and identification of Brucella sppClin. Lab.200349487505
- Al DahoukSTomasoHNocklerKNeubauerHFrangoulidisDLaboratory-based diagnosis of brucellosis--a review of the literature. Part II: serological tests for brucellosisClin. Lab.200349577589
- BrickerBJHallingSMDifferentiation of Brucella abortus bv. 1, 2, and 4, Brucella melitensis, Brucella ovis, and Brucella suis bv. 1 by PCRJ. Clin. Microbiol.19943226602666264138
- MatopeGBhebheEMumaJBSkjerveEDjonneBCharacterization of some Brucella species from Zimbabwe by biochemical profiling and AMOS-PCRBMC Res. Notes2009210.1186/1756-0500-2-2612807434
- Al DahoukSEvaluation of Brucella MLVA typing for human brucellosisJ. Microbiol. Methods20076913714510.1016/j.mimet.2006.12.015
- DornelesEMGenetic stability of Brucella abortus isolates from an outbreak by multiple-locus variable-number tandem repeat analysis (MLVA16)BMC. Microbiol.20141418610.1186/1471-2180-14-1864112982
- Le FlechePEvaluation and selection of tandem repeat loci for a Brucella MLVA typing assayBMC. Microbiol.20066910.1186/1471-2180-6-91513380
- BerlinKKorenSChinCSAssembling large genomes with single-molecule sequencing and locality-sensitive hashingNat. Biotechnol.20153362363010.1038/nbt.3238
- KorenSPhillippyAMOne chromosome, one contig: complete microbial genomes from long-read sequencing and assemblyCurr. Opin. Microbiol.20152311012010.1016/j.mib.2014.11.014
- CaoXWhole-genome sequences of Brucella melitensis Strain QY1, isolated from sheep in Gansu, ChinaGenome Announc.20175pii: e0089617
- KurtzSVersatile and open software for comparing large genomesGenome Biol.2004510.1186/gb-2004-5-2-r12395750
- LiWJaroszewskiLGodzikAClustering of highly homologous sequences to reduce the size of large protein databasesBioinformatics20011728228310.1093/bioinformatics/17.3.282
- NandiTA genomic survey of positive selection in Burkholderia pseudomallei provides insights into the evolution of accidental virulencePLoS. Pathog.20106e100084510.1371/journal.ppat.10008452848565