Abstract
The genus Myroides comprises several species of Gram-negative, non-motile, and non-fermenting bacteria, which have been regarded as non-pathogenic for decades. Multiple recent reports, however, underscore the pathogenic potential that Myroides sp. possesses for humans. These bacteria seem to be resistant to a wide range of antibiotics (including ß-lactams and aminoglycosides). Therefore, treatment options are limited. Knowledge of antimicrobial resistance, however, is based on only one meaningful comprehensive study and on data published from case reports. This lack of data motivated us to test 59 strains from our Myroides collection (43 M. odoratimimus and 16 M. odoratus) for resistance against 20 commonly used antibiotics. We also performed molecular analyses to reveal whether our bacteria harbor the genus-specific M. odoratimimus metallo-ß-lactamase (MUS-1) or the M. odoratus metallo ß-lactamase (TUS-1), and other ß-lactamases, which may provide an explanation for the extended antimicrobial resistance.
These authors contributed equally: Florian Gunzer, Wolfram W. Rudolph.
Introduction
The bacteria known as Myroides today were first described as Bacterium faecale aromaticum by Stutzer in 1923Citation1. Six years later, in 1929, these bacteria were renamed Flavobacterium odoratumCitation2. Finally, in 1996, Vancanneyt et al. undertook a new reclassification based on modern techniques that included DNA–rRNA hybridization, DNA–DNA hybridization, analysis of whole-cell protein patterns, fatty acid composition, and the phenotypeCitation3. Based on their results, they introduced a new genus, Myroides, with the species M. odoratus (formerly F. odoratum) and a new species, M. odoratimimus, into the bacterial taxonomyCitation3. The bacteria are Gram-negative, non-motile, non-fermenting, and rod-shaped. Their natural habitat is soil and waterCitation3–Citation5. Infections caused by Myroides sp. comprise necrotizing fasciitis, soft tissue infections, ventriculitis, pneumonia, and sepsis. Immunocompromised patients are mostly affected, although being immunocompromised does not seem to be absolutely prerequisite to acquiring a Myroides infectionCitation6–Citation12. In addition, two studies on outbreaks have been publishedCitation13,Citation14. Furthermore, the outcome was fatal in two case reportsCitation9,Citation13–Citation15. Since Myroides sp. are resistant to a wide range of antibiotics, including ß-lactams and aminoglycosides, choosing an appropriate empirical antimicrobial therapy is challenging. Moreover, knowledge about the specific resistance profile of Myroides sp. is based on data mainly obtained from case reports and two publicationsCitation16,Citation17. In the present study, we therefore compare minimal inhibitory concentration (MIC) results for 20 important antibiotics against 59 strains from our Myroides collection (43 M. odoratimimus and 16 M. odoratus). The data were evaluated according to the guidelines published by EUCAST in 2017. Furthermore, PCR analysis and whole-genome sequencing approaches were applied to detect corresponding genes of important ß-lactamases of the OXA-types, VIM-types, IMP-types, AmpC-types, KPC-types, and NDM-types and for the Myroides sp.-specific enzymes MUS-1 and TUS-1.
Results
Antimicrobial susceptibility
The susceptibility profile and the MIC distribution of all strains tested are summarized in Tables and . The MIC results of each strain are additionally provided in Table S1. Only a few M. odoratimimus and M. odoratus strains are susceptible to ampicillin (M. odoratimimus, n = 1; M. odoratus, n = 5) and piperacillin/tazobactam (M. odoratimimus, n = 2; M. odoratus, n = 0). Interestingly, more of the tested strains showed intermediate susceptibility to ampicillin (M. odoratimimus, n = 19; M. odoratus, n = 2) than to piperacillin/tazobactam (M. odoratimimus, n = 9; M. odoratus, n = 0). No strain was susceptible to ceftazidime, cefepime, and aztreonam. In total, 4 M. odoratimimus strains and no M. odoratus strain were susceptible to imipenem. In contrast to this observation, 32 M. odoratimimus and 8 M. odoratus strains were susceptible to meropenem. The MIC results of the quinolones—ciprofloxacin, levofloxacin, and moxifloxacin—were also compared in this study. Most strains were susceptible to moxifloxacin (M. odoratimimus, n = 39; M. odoratus, n = 15). In contrast to this, 5 M. odoratimimus strains and 7 M. odoratus strains tested were susceptible to levofloxacin. Only 1 M. odoratimimus strain and 1 M. odoratus strain were susceptible to ciprofloxacin. Of all quinolones tested, moxifloxacin showed the lowest MIC values in this antibiotic class. In addition, only 1 M. odoratimimus strain and 1 M. odoratus were susceptible to tigecycline.
MIC distribution of 43 M. odoratimimus strains
MIC distribution of 16 M. odoratus strains
No breakpoints were available for trimethoprim/sulfamethoxazole, fosfomycin, colistin, gentamicin, amikacin, erythromycin, azithromycin, daptomycin, and rifampicin. The MIC results determined for fosfomycin, colistin, gentamicin, amikacin, and daptomycin are all at a high range. Therefore, antimicrobial resistance to these antibiotics may be assumed. Regarding trimethoprim/sulfamethoxazole, erythromycin, azithromycin, and rifampicin, the MIC results ranged from low to high levels.
Detection of relevant β-lactamase genes
All strains were negative for genes coding for OXA-type, VIM-type, IMP-type, and NDM β-lactamases, and for KPC. The AmpC β-lactamase gene was detected in one isolate (DSM 100899, Table S2).
Homologies of blaMUS-1 and blaTUS-1
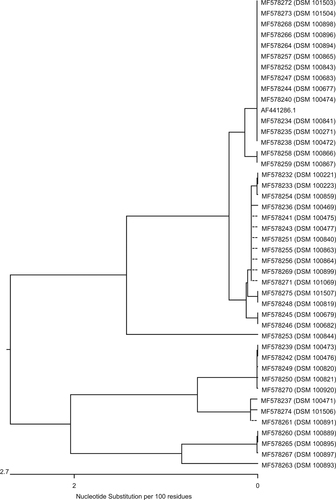
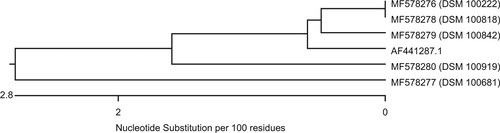
Genes homologous to blaMUS-1 were only detected in M. odoratimimus; genes homologous to blaTUS-1, only in M. odoratus isolates. Figure is a dendrogram that presents the nucleotide sequence similarity of all genes of our M. odoratimimus strains that show homology to the carbapenemase MUS-1 gene of M. odoratimimus CIP 103073 (GenBank accession #AF441286.1). Figure is also a dendrogram. It shows the nucleotide sequence similarities of our M. odoratus strains compared to the carbapenemase TUS-1 gene of M. odoratus CIP 103105 (GenBank accession #AF441287.1) in the same fashion. No DNA sequence homologies to blaTUS-1 were found in 11 out of 16 isolates (Table S3).
Comparison of nucleotide and protein sequences
Nucleotide sequences were transferred into protein sequences using Lasergene software version 14.0.10 (DNASTAR, Madison, WI, USA). Alignments of all MUS-1 and all TUS-1 genes, as well as their protein translations, are illustrated in Figs. S1– 4. When the sequences are compared, alterations are only observed at certain positions in the genes homologous to blaMUS-1 and blaTUS-1. The active centers of both enzymes, however, were not involved. This observation is confirmed by the translated protein sequences. The maximum divergence between the sequences of genes homologous to blaMUS-1 was 5.6% in the nucleotide composition and 6.5% in the amino acid (AA) composition. Additionally, the maximum divergence of genes homologous to blaTUS-1 was 5.1% at the nucleotide level and 2.8% at the AA level.
Discussion
Bacteria of the genus Myroides are rare human pathogens that cause severe infections (including blood stream infections, necrotizing fasciitis, ventriculitis, and pneumonia) and affect mainly immunocompromised patientsCitation6,Citation7,Citation9,Citation18,Citation19. Most clinical case reports focus on M. odoratimimus and M. odoratus. Until now, whether other species of the genus Myroides, such as M. pelagicus, M. profundi, M. marinus, M. phaeus, M. guanonis, M. xuanwuensis, M. injenensis, and M. indicus, are pathogenic for humans has been uncertainCitation20–Citation27. M. odoratimimus and M. odoratus are described to be resistant to a large number of antibiotics. The available antimicrobial susceptibility data on Myroides sp. are mostly based on case reports and four systematic studiesCitation16,Citation28,Citation29. Two of these studies included only a few isolatesCitation28,Citation29 and therefore are limited. Holmes et al. tested 28 F. odoratum isolates against 15 antibioticsCitation16, whereas Hu et al. summarized and compared data published in the literature and added data that previously were available only in ChinaCitation17. However, those authors did not distinguish between M. odoratimimus and M. odoratus. As far as we know, this is the only systematic study that provides reliable susceptibility data. Because F. odoratum was reclassified and divided into the two new species—M. odoratimimus and M. odoratus—approximately 20 years after Holmes and coworkers published their dataCitation3,Citation16, resistance analysis of these two Myroides species must be repeated. Here, we provide susceptibility data for both M. odoratus and M. odoratimimus for the first time. Although all strains included in the study by Holmes et al. were resistant to ampicillinCitation16, we found that 2.3% (n = 1) of the M. odoratimimus and 31.3% (n = 5) of the M. odoratus strains were susceptible to ampicillin (Tables and ). Additionally, 4.7% (n = 2) of our M. odoratimimus strains but none of the M. odoratus strains (Tables and ) were susceptible to piperacillin/tazobactam. Myroides sp. can show any level of susceptibility, including resistance to piperacillin/tazobactamCitation6,Citation7,Citation9,Citation11. However, none of our strains were susceptible to ceftazidime and cefepime (Tables and ). Our finding is also in accordance with previous reportsCitation6,Citation11,Citation15. Only Crum-Cianflone et al. reported on an M. odoratus isolate with intermediate susceptibility to cefepimeCitation9. Our data and data from previous studies show that Myroides sp. are resistant to aztreonamCitation6,Citation7,Citation9,Citation11 (Tables and ). We found that 83.7% (n = 36) of our M. odoratimimus strains and all M. odoratus strains were resistant to imipenem, which is consistent with data from other studies showing susceptibilityCitation6, intermediate resistanceCitation9, or resistanceCitation7,Citation15. We observed that 74.4% (n = 32) of our M. odoratimimus strains and 50.0% (n = 8) of the M. odoratus strains are susceptible to meropenem (Tables and ), which has also been reported by othersCitation6,Citation9. Therefore, meropenem may be more useful for the treatment of Myroides infections than imipenem. While investigating quinolones, we found a gradual change in susceptibility. Most strains were resistant to ciprofloxacin (Tables and ): 83.7% (n = 36) of the M. odoratimimus strains and 68.7% (n = 11) of the M. odoratus strains were resistant, whereas only 2.3% (n = 1) of the M. odoratimimus strains and 6.3% (n = 1) of the M. odoratus strains were susceptible. However, resistant, as well as susceptible, isolates have been reported beforeCitation6,Citation7,Citation9,Citation18. A larger number of isolates were susceptible to levofloxacin compared to ciprofloxacin (Tables and ), namely, 11.6% (n = 5) of the M. odoratimimus and 43.8% (n = 7) of the M. odoratus strains. Interestingly, moxifloxacin testing revealed significantly lower MIC values compared to levofloxacin and ciprofloxacin (Tables and ). In fact, only one M. odoratus isolate was resistant, but 90.7% (n = 39) of our M. odoratimimus isolates and 93.8% (n = 15) of our M. odoratus isolates were susceptible (Table ). Based on these in vitro data, moxifloxacin may therefore be the best quinolone to be used. Ali et al. recently reported the successful use of moxifloxacin in a patient with canaliculitisCitation30. To our knowledge, susceptibility testing for tigecycline on Myroides sp. has not been published before, and our data on tigecycline show that most strains of both M.odoratimimus and M. odoratus are resistant (Tables and ). No EUCAST guidelines exist for trimethoprim/sulfamethoxazole, fosfomycin, colistin, amikacin, erythromycin, azithromycin, daptomycin, and rifampicin. Nevertheless, we could detect very high MIC values for colistin, fosfomycin, and daptomycin for all our isolates. For this reason, M. odoratimimus and M. odoratus may be assumed to be naturally resistant to these antibiotics. Judging from the high MIC values, our strains seem to be naturally resistant against the aminoglycosides gentamicin and amikacin. Our findings are in accordance with the data reported from several laboratories working on MyroidesCitation7,Citation9,Citation11,Citation15,Citation16. The MIC values for trimethoprim/sulfamethoxazole span a wide range (for M. odoratimimus 0.125–8 µg/ml and for M. odoratus 0.5–4 µg/ml). Using different breakpoints for their antimicrobial resistance testing, Holmes et al. found both susceptible and resistant strains to trimethoprim/sulfamethoxazoleCitation16. Similarly, a wide range of MIC values was found for the macrolide antibiotics erythromycin and azithromycin and for rifampicin.
Very little is known about the resistance mechanisms in Myroides today. In 2002, Mammeri et al. identified two ß-lactamases in Myroides sp.—MUS-1 and TUS-1—and these metalloenzymes share 73% AA identityCitation31. Based on sequence analyses, both MUS-1 and TUS-1 could be identified as members of the B1 subclass family according to the classification by AmblerCitation32, which has been expanded by Galleni et alCitation33. According to the kinetic studies performed by Mammeri et al., MUS-1 and TUS-1 hydrolyze all ß-lactam antibiotics except aztreonamCitation31. This ability is characteristic of enzymes belonging to the subclass 3a of metallo-ß-lactamases according to the revised functional classification proposed by Bush and Jacoby in 2010Citation34. In contrast, our strains were mostly resistant to aztreonam, and many were susceptible to meropenem (Tables and ). However, meropenem is characteristically hydrolyzed by group 3a ß-lactamasesCitation34. For this reason (resistance against aztreonam and susceptibility to meropenem), we assume that neither MUS-1 nor TUS-1 is expressed at high quantities. Therefore, additional, yet unresolved mechanisms, such as porin-mutations or other ß-lactamase hydrolyzing enzymes, likely are present and explain the extended resistance of both M. odoratimimus and M. odoratus against ß-lactam antibiotics. This observation is also supported by the fact that 11 of the M. odoratus strains do not contain the TUS-1 genes. However, all our M. odoratimimus possess MUS-1. Nevertheless, all strains of M. odoratus, irrespective of the presence or absence of the blaTUS-1 gene, show similar resistance profiles (Table S1). A Myroides strain, assigned to M. odoratimimus based on 16S rDNA sequencing, without blaMUS-1 has previously been reported by Dharne et alCitation35. Yet this isolate was reported to be highly resistant to β-lactam antibioticsCitation35. Based on molecular and phenotypic analyses, Al-Bayssari et al. proposed a new variant of a MUS ß-lactamase in 2015, which was named MUS-2 and possessed 98.78% AA homology to MUS-1Citation36. However, in contrast to the work by Mammeri et al.Citation31,Citation36, these authors did not perform an in-depth analysis of the enzyme kinetics.
A comparison of the genomic data from all 48 Myroides strains carrying blaMUS-1/blaTUS-1 to the sequences published by Mammeri et al.Citation31,Citation36 show that the encoding genes have a fairly high homology, ranging from 94.7 to 100% for blaMUS-1 and from 94.9 to 99.3% for blaTUS-1 (Table S4 and Figs. S1, S3). At the protein level, MUS-1 shares 93.5–100% homology and TUS-1 97.2–99.6% (Table S4 and Figs. S2, S4). Notably, all 43 M. odoratimimus strains harbor a MUS-1 encoding gene, whereas, of the 16 M. odoratus isolates, 11 were negative for blaTUS-1. Furthermore, different clusters of “MUS” or “TUS” genes can be defined (Figs. ). Only 30.2% (n = 13) of our M. odoratimimus strains harbor genes that are genetically identical to blaMUS-1. We could not detect a gene that was 100% identical to the originally described nucleotide sequence for blaTUS-1. In other words, the variability of M. odoratus with respect to the gene blaTUS-1 is large. Only five genes were homologous to the sequence initially described by Mammeri et al.Citation31,Citation36, with variation ranging from 94.9 to 99.3%. Comparison of the nucleotide and AA sequences, however, suggests that genomic differences may not have an effect on the active center of the enzymes, and therefore may not affect the hydrolytic activity. Additional data from a similar comparative study on enzyme kinetics, however, would elucidate whether these sequences encode for new variants. Therefore, the results and the conclusions drawn by Al-Bayssari et al.Citation36 regarding a newly described MUS-2 have to be corroborated. Notably, our M. odoratimimus isolate DSM 100899 harbors the gene encoding the ß-lactamase AmpC. It can be located chromosomally or on specific resistance (R-) plasmidsCitation37. We believe this is the first report of an AmpC encoding gene detected in Myroides sp. Furthermore, the R-plasmids seem to play an important role in antimicrobial resistance mechanisms in Myroides sp. Kono et al., for instance, reported on resistance genes against ampicillin, carbenicillin, and erythromycin which were located on an R-plasmidCitation38. In addition, Kuai et al. reported on a KPC-2-positive F. odoratum strain in 2011Citation39. In general, a variety of genes for antimicrobial resistance seems to be characteristic for Myroides sp. Elucidating the exact mechanisms of antimicrobial resistance, however, is a matter of ongoing and future research.
Materials and methods
Bacterial isolates
A collection of clinical isolates of Myroides strains was created over the course of 4 years (Table S5). The strains were collected during the routine diagnostics at the Institute for Medical Microbiology and Hygiene, TU Dresden, Germany. Additional strains were provided by Bodo R. Eing and Sebastian Bertram (Synlab Medizinisches Versorgungszentrum Augsburg, Augsburg, Germany). A few isolates were of veterinary origin (Table S5). The bacteria were confirmed as M. odoratimimus or M. odoratus by two independent methods: 16S rRNA gene sequencing and MALDI-TOF MS (Bruker Daltonik, Bremen, Germany). We have addressed the suitability of this identification strategy, as well as the performance and reliability of both methods, in previous studiesCitation40,Citation41. In the present investigation, we included 59 strains, with 43 being identified as M. odoratimimus and 16 as M. odoratus. All strains were also deposited in the “Open Collection” of the Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany).
Antimicrobial susceptibility testing
The MIC tests were performed, and the results were evaluated by applying the guidelines for PK/PD (non-species related) breakpoints according to the criteria published by EUCAST (the European Committee on Antimicrobial Susceptibility Testing). EUCAST breakpoint tables were created for the interpretation of the MICs and zone diameters using Version 7.1, 2017 (http://www.eucast.org/clinical_breakpoints/). In brief, a McFarland standard of 0.5 was created for each bacterium using NaCl and a DensiCHEK densitometer (bioMérieux, Nürtingen, Germany). The suspended bacteria were plated with the help of a cotton swab on Müller-Hinton Agar (Oxoid Deutschland, Wesel, Germany). Then, E-Test strips (bestbion, Cologne, Germany) for each antibiotic were placed on the agar plates. The plates were incubated for 18 ± 2 h at 37 °C and 5% CO2. Then, the MIC results were determined. The ATCC strains Escherichia coli ATCC® 25922™, Pseudomonas aeruginosa ATCC® 27853™, and Staphylococcus aureus ATCC® 25923™ served as the controls. All E-Test strips used in this study and their MIC ranges are listed in Table S6.
Real-time PCR analysis for β-lactamase gene detection
All samples were cultivated on blood agar plates (25 °C, 24 h), and DNA was extracted with a QB-EX-50 QuickBlue extraction kit using magnetic nanoparticles (Q-Bioanalytic, Bremerhaven, Germany). Real-time PCR analysis was carried out on all isolates to evaluate the prevalence of the blaOXA, blaVIM, blaIMP, blaAmpC, and blaKPC carbapenemase genes and the blaNDM metallo-ß-lactamase gene. QuickBlue RealQuick PCR master mix—SYBR Green (QB-RT-ESBL-50-SYBR, Q-Bioanalytic) for carbapenemase gene detection was prepared in 15 µl of master mix that included Taq polymerase, buffer, dNTPs, the specific primer set, and SYBR Green. The amplification and detection of the metallo-ß-lactamase blaNDM amplicon was carried out using QuickBlue RealQuick PCR master mix (QB-RT-ESBL-50, Q-Bioanalytic), which included Taq polymerase, buffer, dNTPs, specific primer set, and probe (Fam-labeled). Two microliters of DNA template (5–10 ng genomic DNA) was added to each PCR application, bringing the total volume to 17 µl. Amplification reactions were carried out using a real-time thermal cycler (LightCycler 480 II, Roche Deutschland, Grenzach-Wyhlen, Germany) with the following program: one cycle of 5 min at 95 °C, 35 cycles with denaturation at 95 °C for 15 s and annealing at 60 °C for 20 s; and an extension at 72 °C for 15 s. Data were analyzed using the Roche LightCycler Software 1.5. Primer sets were designed with Integrated DNA Technologies “PrimerQuest Tool” (https://eu.idtdna.com/PrimerQuest/Home/Index) using genomic sequences from NCBI GenBank. Multiple alignments from several ß-lactamase genes were analyzed with the Clustal Omega program (https://www.ebi.ac.uk/Tools/msa/clustalo/), and the primer sets were optimized to ensure detection of a broad range of bacterial resistance genes. Klebsiella pneumoniae NCTC 13443 (blaNDM), K. pneumoniae NCTC 13440 (blaVIM-1), K. pneumoniae NCTC 13442 (blaOXA-48), K. pneumoniae BAA 1705 (blaKPC), E. coli NCTC 13476 (blaIMP), and Enterobacter cloacae BAA 1143 (blaAmpC) strains were used as negative and positive controls. The results are illustrated in Table S2.
Detection of MUS-1 and TUS-1 encoding genes using a whole-genome sequencing approach
Libraries for whole-genome sequencing were prepared with Nextera XT DNA Library Prep Kit (Illumina, San Diego, CA, USA). Sequencing was performed on the Illumina HiSeq 2500 (Illumina) for 100 cycles in both directions at the Helmholtz Centre for Infection Research (Braunschweig, Germany). Hereby, coverages of approximately 100× were obtained for each of the isolates. Assembly of short read genome data was performed using Velvet 1.2.10Citation42 with a kmer size of 61 for all isolates. Resulting contig sets were screened for known antibiotic resistance genes querying the ResFinder 2.1 databaseCitation43 in an automated approach. Hereby, minimum values of 60% query coverage and 90% identity have been used as search parameters. A comparative analysis of the DNA sequences obtained was performed using the Molecular Evolutionary Genetics Analysis software package version 7Citation44. Protein sequences were created using DNASTAR Lasergene 14.0.10. (DNASTAR). Average nucleotide identity (ANI) values have been calculated on the whole-genome level using the software FastANI (https://github.com/ParBLiSS/FastANI) (Table S3). Therefore, all M. odoratimimus strains used in this study were compared with M. odoratimimus CCUG39352T (draft assembly ASM148541v1, https://www.ncbi.nlm.nih.gov/assembly/GCF_001485415.1), and all M. odoratus strains were compared with M. odoratus DSM 2801T (GenBank accession #CM001437).
Table S1 (DOCX 110 kb)
Download MS Word (46.8 KB)Table S2 (DOCX 73 kb)
Download MS Word (30.5 KB)Table S3 (DOCX 82 kb)
Download MS Word (33.4 KB)Table S4 (DOCX 68 kb)
Download MS Word (29.2 KB)Table S5(DOCX 27 kb)
Download MS Word (27.9 KB)Table S6 (DOCX 34 kb)
Download MS Word (16.3 KB)Figure S1(PNG 767 kb)
Download PNG Image (767.3 KB)Figure S2(PNG 479 kb)
Download PNG Image (479.4 KB)Figure S3(PNG 118 kb)
Download PNG Image (118.6 KB)Figure S4(PNG 74 kb)
Download PNG Image (74.6 KB)Acknowledgements
This work was supported by “Zeidler-Forschungs-Stiftung” (grant to P.S.). We thank Jörg Overmann for genome sequencing and scientific support, Enno Jacobs for his continuing support, and Sigrid Gäbler and Franziska Klann for expert technical assistance.
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41426-018-0061-x).
References
- StutzerMJZur Frage Über die Fäulnisbakterien im DarmZentralbl Bakteriol Parasitenkd Infektionskr Hyg Abt I Orig192391 87 90
- StutzerMKwaschninaAAussaaten aus dem Fäzes des Menschen gelbe Kolonien bildende Bakterien (Gattung Flavobacterium u. a.)Zentralbl Bakteriol Parasitenkd Infektionskr Hyg Abt I Orig1929113219225
- VancanneytMReclassification of Flavobacterium odoratum (Stutzer 1929) strains to a new genus, Myroides, as Myroides odoratus comb. nov. and Myroides odoratimimus sp. novInt. J. Syst. Evol. Microbiol.199646926932
- InbakandanDMurthyPSVenkatesanRKhanSA16S rDNA sequence analysis of culturable marine biofilm forming bacteria from a ship’s hullBiofouling20102689389910.1080/08927014.2010.530347
- JinFGenetic diversity and phylogeny of antagonistic bacteria against Phytophthora nicotianae isolated from tobacco rhizosphereInt. J. Mol. Sci.2011123055307110.3390/ijms120530553116175
- BenedettiPRassuMPavanGSeftonAPellizzerGSeptic shock, pneumonia, and soft tissue infection due to Myroides odoratimimus: report of a case and review of Myroides infectionsInfection20113916116510.1007/s15010-010-0077-1
- GreenBTGreenKNolanPEMyroides odoratus cellulitis and bacteremia: case report and reviewScand. J. Infect. Dis.20013393293410.1080/00365540110077065
- HsuehPRWuJJHsiueTRHsiehWCBacteremic necrotizing fasciitis due to Flavobacterium odoratumClin. Infect. Dis.1995211337133810.1093/clinids/21.5.1337
- Crum-CianfloneNFMatsonRWBallon-LandaGFatal case of necrotizing fasciitis due to Myroides odoratusInfection20144293193510.1007/s15010-014-0626-0
- DeepaRVenkateshKGParveenJDBanuSTJayalakshmiGMyroides odoratus and Chryseobacterium indologenes: two rare isolates in the immunocompromisedIndian J. Med. Microbiol.20143232733010.4103/0255-0857.136592
- MarakiSSarchianakiEBarbagadakisSMyroides odoratimimus soft tissue infection in an immunocompetent child following a pig bite: case report and literature reviewBraz. J. Infect. Dis.20121639039210.1016/j.bjid.2012.06.004
- MacfarlaneDEBaum-ThureenPCrandonIFlavobacterium odoratum ventriculitis treated with intraventricular cefotaximeJ. Infect.19851123323810.1016/S0163-4453(85)93228-1
- KtariSNosocomial outbreak of Myroides odoratimimus urinary tract infection in a Tunisian hospitalJ. Hosp. Infect.201280778110.1016/j.jhin.2011.09.010
- YagciAMolecular typing of Myroides odoratimimus (Flavobacterium odoratum) urinary tract infections in a Turkish hospitalEur. J. Clin. Microbiol. Infect. Dis.20001973173210.1007/s100960070001
- PrateekSGuptaPMittalGSinghAKFatal case of pericardial effusion due to Myroides odoratus: a rare case reportJ. Clin. Diagn. Res.20159DD01DD024668411
- HolmesBSnellJJLapageSPFlavobacterium odoratum: a species resistant to a wide range of antimicrobial agentsJ. Clin. Pathol.197932737710.1136/jcp.32.1.731145571
- HuSAntibiotic resistance mechanisms of Myroides spJ. Zhejiang Univ. Sci. B20161718819910.1631/jzus.B15000684794510
- BachmeyerCCellulitis due to Myroides odoratimimus in a patient with alcoholic cirrhosisClin. Exp. Dermatol.2008339798
- Endicott-YazdaniTRDhimanNBenavidesRSpakCWMyroides odoratimimus bacteremia in a diabetic patientProc. Bayl. Univ. Med. Cent.20152834234310.1080/08998280.2015.119292684462216
- YoonJManeeratSKawaiFYokotaAMyroides pelagicus sp. nov., isolated from seawater in ThailandInt. J. Syst. Evol. Microbiol.2006561917192010.1099/ijs.0.64336-0
- ZhangXYMyroides profundi sp. nov., isolated from deep-sea sediment of the southern Okinawa TroughFEMS Microbiol. Lett.200828710811210.1111/j.1574-6968.2008.01299.x
- ChoSHChaeSHImWTKimSBMyroides marinus sp. nov., a member of the family Flavobacteriaceae, isolated from seawaterInt. J. Syst. Evol. Microbiol.20116193894110.1099/ijs.0.024067-0
- YanSZhaoNZhangXHMyroides phaeus sp. nov., isolated from human saliva, and emended descriptions of the genus Myroides and the species Myroides profundi Zhang et al. 2009 and Myroides marinus Cho et al. 2011Int. J. Syst. Evol. Microbiol.20126277077510.1099/ijs.0.029215-0
- TomovaAMyroides guanonis sp. nov., isolated from prehistoric paintingsInt. J. Syst. Evol. Microbiol.2013634266427010.1099/ijs.0.050831-0
- ZhangZDHeLYHuangZShengXFMyroides xuanwuensis sp. nov., a mineral-weathering bacterium isolated from forest soilInt. J. Syst. Evol. Microbiol.20146462162410.1099/ijs.0.056739-0
- PaekJMyroides injenensis sp. nov., a new member isolated from human urineAntonie van Leeuwenhoek201510720120710.1007/s10482-014-0317-y
- RamHMyroides indicus sp. nov., isolated from garden soilInt. J. Syst. Evol. Microbiol.2015654008401210.1099/ijsem.0.000530
- FraserSLJorgensenJHReappraisal of the antimicrobial susceptibilities of Chryseobacterium and Flavobacterium species and methods for reliable susceptibility testingAntimicrob. Agents Chemother.19974127382741164199
- ChangJCAntimicrobial susceptibility of flavobacteria as determined by agar dilution and disk diffusion methodsAntimicrob. Agents Chemother.19974113011306163904
- AliMJJosephJSharmaSNaikMNCanaliculitis with isolation of Myroides speciesOphthal. Plast. Reconstr. Surg.201533S24S2510.1097/IOP.0000000000000604
- MammeriHBellaisSNordmannPChromosome-encoded beta-lactamases TUS-1 and MUS-1 from Myroides odoratus and Myroides odoratimimus (formerly Flavobacterium odoratum), new members of the lineage of molecular subclass B1 metalloenzymesAntimicrob. Agents Chemother.2002463561356710.1128/AAC.46.11.3561-3567.2002128705
- AmblerRPThe structure of beta-lactamasesPhilos. Trans. R. Soc. Lond. B Biol. Sci.198028932133110.1098/rstb.1980.0049
- GalleniMStandard numbering scheme for class B beta-lactamasesAntimicrob. Agents Chemother.20014566066310.1128/AAC.45.3.660-663.200190352
- BushKJacobyGAUpdated functional classification of beta-lactamasesAntimicrob. Agents Chemother.20105496997610.1128/AAC.01009-09
- DharneMSAntibacterial activities of multi drug resistant Myroides odoratimimus bacteria isolated from adult flesh flies (Diptera: Sarcophagidae) are independent of metallo beta-lactamase geneBraz. J. Microbiol.20083939740410.1590/S1517-838220080002000353768384
- Al-BayssariCGuptaSKDabboussiFHamzeMRolainJMMUS-2, a novel variant of the chromosome-encoded beta-lactamase MUS-1, from Myroides odoratimimusNew Microbes New Infect.20157677110.1016/j.nmni.2015.06.0074522612
- BebroneCCurrent challenges in antimicrobial chemotherapy: focus on ss-lactamase inhibitionDrugs20107065167910.2165/11318430-000000000-00000
- KonoMSasatsuMMakinoKR-plasmid conferring resistance to ampicillin, carbenicillin and erythromycin in a Flavobacterium odoratum strainMicro. Lett.1980145558
- KuaiSHuangLPeiHChenYLiuJImipenem resistance due to class A carbapenemase KPC-2 in a Flavobacterium odoratum isolateJ. Med. Microbiol.2011601408140910.1099/jmm.0.029660-0
- SchröttnerPRudolphWWEingBRBertramSGunzerFComparison of VITEK2, MALDI-TOF MS, and 16S rDNA sequencing for identification of Myroides odoratus and Myroides odoratimimusDiagn. Microbiol. Infect. Dis.20147915515910.1016/j.diagmicrobio.2014.02.002
- SchröttnerPGunzerFSchüppelJRudolphWWIdentification of rare bacterial pathogens by 16S rRNA gene sequencing and MALDI-TOF MSJ. Vis. Exp.2016113e53176
- ZerbinoDRBirneyEVelvet: algorithms for de novo short read assembly using de Bruijn graphsGenome Res.20081882182910.1101/gr.074492.1072336801
- ZankariEIdentification of acquired antimicrobial resistance genesJ. Antimicrob. Chemother.2012672640264410.1093/jac/dks2613468078
- KumarSStecherGTamuraKMEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasetsMol. Biol. Evol.2016331870187410.1093/molbev/msw054