Abstract
Carbapenem-resistant Acinetobacter baumannii is the top-ranked pathogen in the World Health Organization priority list of antibiotic-resistant bacteria. It emerged as a global pathogen due to the successful expansion of a few epidemic lineages, or international clones (ICs), producing acquired class D carbapenemases (OXA-type). During the past decade, however, reports regarding IC-I isolates in Latin America are scarce and are non-existent for IC-II and IC-III isolates. This study evaluates the molecular mechanisms of carbapenem resistance and the epidemiology of 80 non-duplicate clinical samples of A. baumannii collected from February 2014 through April 2016 at two tertiary care hospitals in Lima. Almost all isolates were carbapenem-resistant (97.5%), and susceptibility only remained high for colistin (95%). Pulsed-field gel electrophoresis showed two main clusters spread between both hospitals: cluster D containing 51 isolates (63.8%) associated with sequence type 2 (ST2) and carrying OXA-72, and cluster F containing 13 isolates (16.3%) associated with ST79 and also carrying OXA-72. ST2 and ST79 were endemic in at least one of the hospitals. ST1 and ST3 OXA-23-producing isolates were also identified. They accounted for sporadic hospital isolates. Interestingly, two isolates carried the novel OXA-253 variant of OXA-143 together with an upstream novel insertion sequence (ISAba47). While the predominant A. baumannii lineages in Latin America are linked to ST79, ST25, ST15, and ST1 producing OXA-23 enzymes, we report the emergence of highly resistant ST2 (IC-II) isolates in Peru producing OXA-72 and the first identification of ST3 isolates (IC-III) in Latin America, both considered a serious threat to public health worldwide.
These authors contributed equally: Saúl Levy-Blitchtein, Ignasi Roca.
Introduction
Acinetobacter baumannii is an opportunistic nosocomial pathogen responsible for a broad range of nosocomial infectionsCitation1, including ventilator-associated pneumonia and bacteremia (35–52% mortality)Citation1,Citation2, as well as skin and soft tissue infections, endocarditis, urinary tract infections, and meningitisCitation1,Citation3. Nosocomial isolates of this bacterium are often resistant to most currently available antibiotics. Carbapenem-resistant A. baumannii has recently been considered the most critical pathogen for public health, topping the global priority list of antibiotic-resistant bacteria published by the World Health OrganizationCitation4. Acinetobacter baumannii strains can develop resistance to all the antibiotics availableCitation5. Outbreaks caused by multidrug-resistant (MDR), extensively drug-resistant (XDR), and even pan-drug-resistant (PDR) strains having been reported worldwideCitation6–Citation9. In the past decade, resistance rates have been rising, and recent reports show a steady increase in carbapenem resistance among A. baumannii strainsCitation8,Citation10–Citation13.
Resistance to carbapenems in A. baumannii is usually mediated by the expression of carbapenem-hydrolyzing class D β-lactamases, also known as OXA-type carbapenemases, although the expression of different class B metallo-β-lactamases (MBLs), or even Klebsiella pneumoniae carbapenemase (KPC) enzymes, has also been reportedCitation5. The OXA-type enzymes described in A. baumannii belong to six different families, namely the intrinsic OXA-51 oxacillinase family, which is usually chromosomally encoded but has rarely been reported in plasmidsCitation14, and the acquired OXA-23, OXA-24, OXA-58, OXA-143, and OXA-235 familiesCitation5. Although the population structure of A. baumannii strains is quite diverse, there seems to be a clonal spread of a few epidemic lineages that predominate over the restCitation15. In particular, the international clones I–III account for most A. baumannii infections worldwide and are usually associated with the production of OXA-23-like, OXA-24-like, or OXA-58-like enzymesCitation15.
Our knowledge of the epidemiology and antibacterial susceptibility profiles of A. baumannii, however, is still incomplete in many parts of the world, including many countries in Latin America. The present study was designed to evaluate the phenotypic resistance patterns, the presence of carbapenem resistance mechanisms, and the clonal relatedness of A. baumannii isolates circulating in Lima, Peru.
Results
Bacterial isolates
A total of 80 non-redundant A. baumannii isolates were recovered from blood (n = 59, 73.8%), bronchial aspirate (n = 15, 18.8%), soft tissue (n = 2, 2.5%), cerebrospinal fluid (n = 2, 2.5%), and urine samples (n = 2, 2.5%) of different patients admitted to two tertiary care hospitals in Lima. Fifty-three isolates were recovered at the Instituto Nacional de Enfermedades Neoplásicas (INEN) from February 2014 through April 2016, and the remaining 27 isolates originated from inpatients at the Hospital Nacional Arzobispo Loayza (HNAL) from July through October 2015.
Antibiotic susceptibility
Overall, antimicrobial susceptibility testing by disc diffusion reported high resistance rates to most of the antibiotics tested. More specifically, resistance to both imipenem and meropenem was as high as 97.5%, with only two isolates being susceptible to carbapenems, one from each hospital. The non-susceptibility rates to ceftazidime, cefotaxime, piperacillin/tazobactam, and levofloxacin were 98.8%, 97.5% to cefepime, 93.8% to trimethoprim-sulfamethoxazole, 80% to tetracycline, 76.3% to gentamicin, 63.8% to ampicillin/sulbactam, 62.5% to doxycycline, 61.3% to amikacin, and 57.5% to tobramycin. Susceptibility only remained high for colistin (95%); the colistin minimal inhibitory concentrations (MICs) ranged from 0.25 to 16 mg/L. The colistin MIC50 and MIC90 were 1 and 2 mg/L, respectively.
All the isolates were resistant to at least one antibiotic from three different classes. One isolate (1.2%) was resistant to all the antimicrobial agents tested and was therefore classified as PDR. Thirty-seven isolates (46.3%) presented an XDR phenotype, most being susceptible to colistin alone, and the remaining 42 isolates (52.5%) were therefore considered MDR.
Antibiotic susceptibility testing by gradient diffusion of selected strains (see below) showed elevated MIC values of carbapenems (>32 mg/L) in all the strains producing acquired class D carbapenemases as well as elevated MIC values of cephalosporins, aminoglycosides, and quinolones, in good agreement with previous data obtained by disc diffusion (Table ). Interestingly, three strains presented MIC values of tigecycline of 4 mg/L and therefore should be considered resistant according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints for EnterobacteriaceaeCitation16.
Antimicrobial susceptibility and molecular characterization of selected A. baumannii isolates from two tertiary hospitals in Lima, Peru
Detection of carbapenem resistance genes
The presence of genes encoding both intrinsic and acquired class D carbapenemases, KPC, and MBLs was investigated by polymerase chain reaction (PCR). None of the isolates were positive for the genes encoding KPC or MBLs, but the intrinsic blaOXA-51-like gene was detected in all the samples. In two isolates, this gene was the only carbapenemase detected, although it could not be associated with the presence of an upstream ISAba1 element, in good agreement with the carbapenem-susceptible phenotype of these two isolates.
The blaOXA-24-like gene, however, was predominant and present in 65 isolates (81.3%) collected from both hospitals, whereas 11 isolates (13.8%) carried the blaOXA-23-like gene, and 2 isolates (2.5%) proved positive in the multiplex PCR for the blaOXA-143-like gene. Overall, there were no substantial differences between the two hospitals regarding the proportion of isolates carrying different acquired class D carbapenemases (Table ).
Number and percentages of A. baumannii isolates from each participating center carrying different acquired blaOXA genes
Pulsed-field gel electrophoresis
The clonal relatedness was initially studied by pulsed-field gel electrophoresis (PFGE), as described below, which allowed the classification of all the isolates into nine different clusters, or pulsotypes (A–I) (Fig. ). Two major clusters disseminating between the two healthcare settings were identified: pulsotypes D and F, which contained 51 (63.8%) and 13 (16.3%) isolates, respectively. All the isolates in these two clusters were associated with the carriage of OXA-24-like carbapenemases, except for one isolate within pulsotype F with no acquired carbapenemases that was susceptible to carbapenems. OXA-23-like-producing A. baumannii were mostly grouped in pulsotype G (n = 6, 7.5%), containing isolates recovered from both hospitals, and pulsotype A, with only four isolates (5%) that were recovered from INEN. Pulsotype I contained only two isolates also recovered from INEN, but one isolate carried an OXA-23-like enzyme, while the other carried an OXA-143-like oxacillinase. Pulsotypes B, C, E, and H contained singletons (1.3%) producing OXA-24-like (pulsotypes E and H), OXA-143-like (pulsotype B), or no acquired OXA-like enzymes (pulsotype C), the latter isolate being susceptible to carbapenems. Figure shows the temporal and spatial distributions of all the clones.
Isolates in red were selected as representative of each clonal group. Braces indicate classification to the corresponding international clones I–V (IC). Isolates were included in the same pulsotype if their Dice similarity index was ≥85%. Colored squares indicate production of distinct families of acquired OXA-type enzymes: blue: OXA-23; green: OXA-24; black: OXA-143; magenta: none. INEN Instituto Nacional de Enfermedades Neoplásicas, HNAL Hospital Nacional Arzobispo Loayza, MDR multidrug-resistant, XDR extensively drug-resistant, PDR; pan-drug-resistant
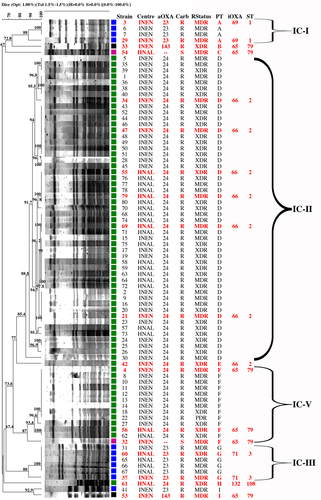
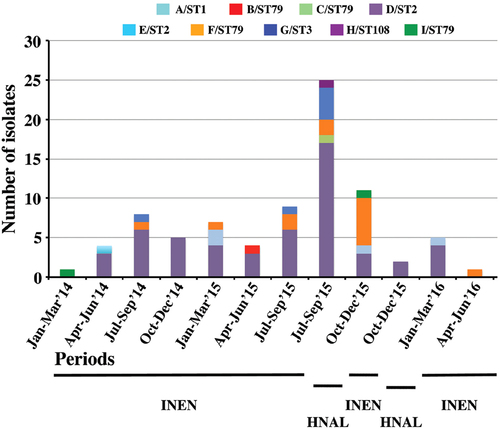
Stacked columns show the number of isolates from each pulsotype/sequence type (PT/ST) recovered at each institute over a 3-month period. INEN Instituto Nacional de Enfermedades Neoplásicas, HNAL Hospital Nacional Arzobispo Loayza
Multilocus sequence typing analysis and OXA sequencing of selected strains
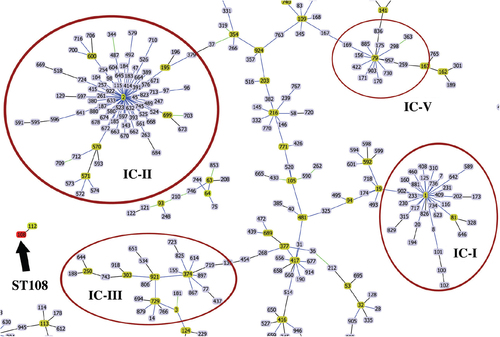
Eighteen isolates were selected for further characterization on the grounds of their clustering in the PFGE dendrogram, susceptibility profiles, and carriage of acquired OXA-type carbapenemases (Table ). Multilocus sequence typing (MLST) analysis using the Pasteur scheme of selected isolates showed full agreement among isolates clustered within the same pulsotype and ST designation (Table ). Isolates from the major PFGE cluster D as well as the singleton from pulsotype E were assigned to ST2 (ST2), which belonged to clonal complex 2 (CC2) and to international clone II. Isolates from the PFGE cluster F, both isolates from pulsotype I and the singletons B and C were assigned to ST79, which was included in CC79, international clone VCitation17. Isolates from pulsotype A were assigned to ST1, belonging to CC1 and international clone I, and those of pulsotype G to ST3, belonging to CC3, international clone III. The H singleton belonged to ST108, which was not assigned to any CC or international clone (Fig. ).
Blue links show SLVs. Founder STs are highlighted in yellow. Reddish circles indicate isolates grouped into a particular international clone (IC). The black arrow points at ST108 and its SLV, ST112
Sequencing of the acquired class D oxacillinase genes in all the selected isolates revealed the presence of the OXA-72 variant in ST2, ST79, and ST108 isolates bearing a blaOXA-24-like gene as well as the presence of the novel OXA-253 variant in the two isolates with a blaOXA-143-like allele. Likewise, sequencing of the intrinsic class D oxacillinase gene identified specific OXA-51-like variants associated with each ST: ST1 isolates presented the blaOXA-69 gene, ST2 was associated with blaOXA-66, ST3 isolates carried the blaOXA-71 gene, ST79 was linked with blaOXA-65, and ST108 had the blaOXA-132 variant (Table ).
Of note, sequence analysis of the genetic structures surrounding the blaOXA alleles identified the presence of the insertion sequence ISAba1 in reverse orientation upstream from the blaOXA-23 gene in all ST1 and ST3 isolates, although we were not able to detect the presence of a downstream ISAba1 copy, suggesting a Tn2008-like structure in these isolatesCitation18. Additionally, a novel insertion sequence was identified in forward orientation upstream of blaOXA-253 in the two strains carrying this OXA enzyme. To provide an attribution number to the new IS, its nucleotide sequence was submitted to ISFinder, the reference center for bacterial insertion sequences (http://www-is.biotoul.fr)Citation19, and it was designated as ISAba47. This novel mobile element showed 82.5% identity at the nucleotide level with ISAba9 and was related to the IS982 family. The downstream region flanking blaOXA-253, however, resembled that previously described by Girlich et alCitation20. A 2334 bp sequence containing the blaOXA-253 gene together with upstream and downstream flanking regions (which included ISAba47) from strain 33 was also submitted to GenBank and can be retrieved under accession number MH347317.
Discussion
In recent decades, we have witnessed the emergence of MDR A. baumannii isolates worldwide, which has been associated with the rapid spread of a few carbapenem-resistant epidemic lineages producing acquired OXA-type carbapenemasesCitation1,Citation15. To date, however, there are only a few studies regarding the epidemiology and carbapenem susceptibility of A. baumannii in Latin America, and this is the first study providing such data from Peru. Interestingly, the predominant A. baumannii lineages reported in Latin America so far are different from those reported in other parts of the world, as international clones II and III are currently absent in this region. International clone II was reported in Brazil in the past, when ST2 isolates producing OXA-23 were described from 1999 through 2003, but it disappeared in 2004Citation21,Citation22. Instead, predominant clones are linked to ST79 (international clone V), ST25 (international clone VII), ST15, and, to a lesser extent, ST1 (international clone I), all of which are mainly associated with the production of OXA-23 enzymesCitation17,Citation23–Citation30.
Nevertheless, there are a few reports describing OXA-72-producing A. baumannii isolates in Latin America, and overall, these isolates are considered less prevalentCitation30–Citation33, or at least they were until recently. In 2017, Pagano et al.Citation34 described the emergence of CC79 and CC15 A. baumannii isolates carrying blaOXA-72 in Brazil and warned of the potential dissemination of these epidemic lineages. Likewise, Nunez Quezada et al.Citation35 reported an outbreak in Ecuador in 2017 caused by OXA-72-producing A. baumanniiCitation35, although the ST of these isolates was not investigated.
The data presented in this study show extremely high resistance rates (>97%) and MIC levels (>32 mg/L) of imipenem and meropenem as well as the predominance of the MDR phenotype among A. baumannii isolates recovered from two tertiary hospitals in the capital city of Peru. Fortunately, the majority (95%) of isolates remain susceptible to colistin, as opposed to several studies reporting increasing rates of colistin-resistant A. baumannii isolates in different countriesCitation8. During the study period, carbapenem resistance in both settings was linked to the widespread dissemination of a major clone producing OXA-72 and belonging to ST2, the founder ST of the epidemic international clone II that has spread globally (Fig. )Citation15. A second group of highly clonal strains producing OXA-72 and belonging to ST79, international clone V, was also identified in both hospitals. In addition, we report the presence of a few sporadic clones producing OXA-23 and linked to international clones I and III (ST1 and ST3, respectively).
International clonal lineages have traditionally been associated with the carriage of specific genetic variants of the intrinsic blaOXA-51 geneCitation24,Citation28,Citation29. This correlation is also shown in our study and supports the genetic relatedness of these isolates with epidemic international lineages. The blaOXA-65, blaOXA-66, blaOXA-69, and blaOXA-71 genes were detected in isolates belonging to ST79, ST2, ST1, and ST3, respectively. We also identified the blaOXA-132 variant in an ST108 isolate that did not belong to any known CC. Interestingly, a single ST108 isolate carrying blaOXA-132 was reported in Lebanon in 2014, and blaOXA-132 was also present in ST15 isolates in Portugal as well as in an ST197 isolate from Saudi ArabiaCitation27,Citation36,Citation37.
The temporal and spatial distributions of the isolates (Fig. ) show that the OXA-72-producing ST2 clone was present in both centers throughout the study period and was likely endemic in INEN, while OXA-72-producing ST79 isolates were sporadic during 2014 and also became endemic at INEN during the second half of 2015. Unfortunately, the collection period of A. baumannii isolates from HNAL was too short to extrapolate endemicity, but as many as 17 ST2 isolates were recovered from July through September 2015, suggesting at least an outbreak situation (Fig. ). Patients are frequently referred between the two hospitals because INEN gathers most patients with neoplastic diseases. That might explain the presence of shared high-risk clones. Of note, 29 out of 37 XDR isolates were OXA-72-producing ST2 isolates. The single PDR isolate, however, was an OXA-72-producing ST79 isolate (Fig. ).
We would also like to highlight the identification of two different ST79 isolates carrying an OXA-253 variant from the OXA-143 family. OXA-253 was first described from an ST79 A. baumannii isolate in Honduras, and to date, only a few additional studies have reported this enzyme in A. baumannii isolates from different STs in BrazilCitation20,Citation38,Citation39. In addition, the genetic environment of blaOXA-253 in this study resembled that described previously by Girlich et al.Citation20, except for the presence of a novel insertion sequence (ISAba47) in the upstream region. ISAba47 might have a role in the mobilization and expression of this gene, as is already the case for the blaOXA-23, blaOXA-58, and blaOXA-235 genes encoding likewise acquired OXA-type carbapenemases that typically present different flanking ISAba sequencesCitation5. To our knowledge, this is the first time that the presence of an IS has been reported to flank a gene belonging to the blaOXA-143 family.
In summary, while the emergence of carbapenem-resistant A. baumannii in Latin America has been associated with the spread of OXA-23-producing ST1, ST15, ST25, and ST79 clonal lineagesCitation28, the results of our study reveal the dissemination of OXA-72-producing XDR clones of A. baumannii in Peru as well as the identification of the epidemic international clones II and III, which were previously absent in this region. The findings presented here are extremely worrisome and should warrant the implementation of infection control measures as well as national and international surveillance measures in the region to contain the further spread of what could be considered nosocomial pandemic lineages of XDR A. baumannii.
Materials and methods
Samples
This study included 80 consecutive clinical isolates of A. baumannii collected from different inpatients at two tertiary care hospitals (HNAL and INEN) from February 2014 through April 2016 in Lima, Peru. Only the first isolate from each patient was included in the study. The samples were obtained from blood, bronchial aspirate, soft tissues, cerebrospinal fluid, and urine, all of which were initially identified as Acinetobacter spp. by the BD Phoenix™ Automated Microbiology System (BD Biosciences, USA) and later identified to the species level by matrix-assisted laser desorption ionization–time of flight mass spectrometry (Bruker, Germany) as described previouslyCitation40.
Antimicrobial susceptibility testing
Antimicrobial susceptibility was assessed by disc diffusion on Mueller–Hinton agar plates in accordance with the Clinical and Laboratory Standards Institute (CLSI) guidelines for the following antimicrobials: ampicillin-sulbactam, piperacillin-tazobactam, cefotaxime, ceftazidime, cefepime, gentamicin, amikacin, levofloxacin, doxycycline, tetracycline, meropenem, imipenem, and trimethoprim-sulfamethoxazole. Susceptibility to colistin was assessed by broth microdilution as recommended by the joint CLSI-EUCAST Polymyxin Breakpoints Working GroupCitation41. The MIC values of selected strains were also determined by gradient diffusion (E test, bioMérieux, Sweden) for the following antimicrobials: imipenem, meropenem, cefotaxime, ceftazidime, cefepime, gentamicin, amikacin, tigecycline, ciprofloxacin, and levofloxacin. The MICs were interpreted according to CLSI clinical breakpoints and expert rules for AcinetobacterCitation41, except for tigecycline, which was interpreted using the EUCAST breakpoints and rules for Enterobacteriaceae (Version 8.0, January 2018)Citation42. Escherichia coli ATCC 25922 and A. baumannii ATCC 19606 were used as quality control strains.
All the isolates were categorized as MDR, XDR, or PDR according to the following ad hoc definitions; MDR, non-susceptible to at least one antimicrobial agent from three classes tested; XDR, non-susceptible to all antimicrobial agents tested but two or fewer; PDR, resistant to all the antimicrobial agents testedCitation7.
Detection of carbapenem resistance genes
The presence of the following carbapenemase-encoding genes was screened by PCR: blaKPC, for serine class A carbapenemases;Citation43blaNDM, blaIMP, blaVIM, blaSPM, and blaSIM for class B MBLs;Citation44 and blaOXA-51-like, blaOXA-23-like, blaOXA-24-like, blaOXA-58-like, blaOXA-143-like, and blaOXA-235-like, for class D oxacillinasesCitation45–Citation47. Amplification products were purified from agarose gels (SpinPrepTM Gel DNA Kit, San Diego, CA, USA) and sent for Sanger sequencing (Macrogen, Korea) whenever necessary. The genetic identity of blaOXA genes was determined upon pairwise sequence alignment with reference sequences retrieved from http://www.lahey.org/Studies/.
The presence of IS sequences flanking the blaOXA genes was studied by PCR using specific primers as well as by inverse PCR and primer walking whenever needed. IS structures were annotated manually using BLASTn and the NCBI bacterial and ISFinder databasesCitation19,Citation48.
Molecular typing
PFGE was performed as described previouslyCitation11, using genomic digestions with the ApaI restriction enzyme and a CHEF-DRIII system (Bio-Rad Laboratories). Molecular patterns were analyzed with InfoQuestTM FP v.5.4 software (Bio-Rad Laboratories) and the unweighted pair group method with arithmetic mean to create dendrograms based on Dice’s similarity coefficient. Using bandwidth tolerance and optimization values set at 1.5 and 1%, respectively, isolates were considered to belong to the same PFGE cluster (pulsotype) if their Dice similarity index was ≥85%Citation49.
MLST was performed using the Pasteur scheme for A. baumanniiCitation50. The allele sequences and STs of selected strains were identified and retrieved from the PubMLST A. baumannii MLST database (http://pubmlst.org/abaumannii/). The population structure of STs was evaluated using the goeBURST software (http://www.phyloviz.net/goeburst/).
Acknowledgements
This study was supported by Cienciactiva of CONCYTEC, contract no. 164-2016-FONDECYT; Planes Nacionales de I+D+i 2008-2011/2013-2016, Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía y Competitividad, Spanish Network for Research in Infectious Diseases (REIPI RD12/0015/0013 and REIPI RD16/0016/0010); the 2017 call for Strategic Action on Health (PI17/01932), co-financed by European Development Regional Fund “A way to achieve Europe” and operative program Intelligent Growth 2014-2020; and grant 2014 SGR 0653 from the Departament d’Universitats, Recerca i Societat de la Informació, of the Generalitat de Catalunya. I.R. was supported by the Department of Health, Generalitat de Catalunya, grant SLT002/16/00349. Part of these data have been presented as a poster communication at the 18th International Congress on Infectious Diseases, 3–4 March, 2018, Buenos Aires, Argentina, and at the XXVIII-European Congress of Clinical Microbiology and Infectious Diseases (ECCMID), Madrid (Spain), 21–24 April, 2018.
Authors’ contributions
Collected the samples: S.L.-B., S.P.-R., W.V.-T., J.V.-P. Conceived and designed the experiments: I.R., L.M., J.d.V.-M., J.V. Performed the experiments: S.L.-B., I.R., S.P.-R., L.M., J.M.-M., M.J.P. Analyzed the data: S.L.-B., I.R., S.P.-R., W.V.-T., J.V.-P., J.M.-M., M.J.P., J.d.V.-M., J.V. Wrote the paper: S.L.-B., I.R., J.d.V.-M., J.V. All authors critically revised the manuscript for intellectual content. All authors read and approved the final manuscript.
Conflict of interest
The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors declare that they have no conflict of interest.
Ethics approval
This study was performed on clinical laboratory isolates. The authors had no contact or interaction with the patients. Personal information of the patients was not collected, to guarantee anonymity and confidentiality. Ethics approval was obtained from the Committee of the Instituto de Investigación Nutricional (IIN), Lima, Peru.
References
- AntunesLCViscaPTownerKJAcinetobacter baumannii: evolution of a global pathogenPathog. Dis.201471 292 30110.1111/2049-632X.12125
- KempfMInvestigation of Acinetobacter baumannii resistance to carbapenems in Marseille hospitals, south of France: a transition from an epidemic to an endemic situationAPMIS2013121647110.1111/j.1600-0463.2012.02935.x
- DijkshoornLNemecASeifertHAn increasing threat in hospitals: multidrug-resistant Acinetobacter baumanniiNat. Rev. Microbiol.2007593995110.1038/nrmicro1789
- Tacconelli, E. et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 10.1016/s1473-3099(17)30753-3 (2017).
- RocaIEspinalPVila-FarresXVilaJThe Acinetobacter baumannii oxymoron: commensal hospital dweller turned pan-drug-resistant menaceFront. Microbiol.2012314810.3389/fmicb.2012.001483333477
- Chen, C. H. et al. Clonal spread of carbapenem-resistant Acinetobacter baumannii across a community hospital and its affiliated long-term care facilities: a cross sectional study. J. Microbiol. Immunol. Infect. 10.1016/j.jmii.2017.08.001 (2017).
- MagiorakosAPMultidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistanceClin. Microbiol. Infect.20121826828110.1111/j.1469-0691.2011.03570.x
- NowakJHigh incidence of pandrug-resistant Acinetobacter baumannii isolates collected from patients with ventilator-associated pneumonia in Greece, Italy and Spain as part of the MagicBullet clinical trialJ. Antimicrob. Chemother.2017723277328210.1093/jac/dkx3225890771
- RiveraGExtensively drug-resistant Acinetobacter baumannii isolated in a university hospital: role of inter-hospital transmissionJ. Infect. Dev. Ctries.201610969910.3855/jidc.6713
- LadavacREmergence of different Acinetobacter baumannii clones in a Croatian hospital and correlation with antibiotic susceptibilityJ. Glob. Antimicrob. Resist.20171021321810.1016/j.jgar.2017.07.001
- UwingabiyeJClonal diversity and detection of carbapenem resistance encoding genes among multidrug-resistant Acinetobacter baumannii isolates recovered from patients and environment in two intensive care units in a Moroccan hospitalAntimicrob. Resist. Infect. Control201769910.1186/s13756-017-0262-45615474
- VianaGFEvolution of antimicrobial resistance of Acinetobacter baumannii in a university hospitalLett. Appl. Microbiol.20115337437810.1111/j.1472-765X.2011.03109.x
- ZarrilliRPournarasSGiannouliMTsakrisAGlobal evolution of multidrug-resistant Acinetobacter baumannii clonal lineagesInt. J. Antimicrob. Agents201341111910.1016/j.ijantimicag.2012.09.008
- ChenTLEmergence and distribution of plasmids bearing the blaOXA-51-like gene with an upstream ISAba1 in carbapenem-resistant Acinetobacter baumannii isolates in TaiwanAntimicrob. Agents Chemother.2010544575458110.1128/AAC.00764-102976157
- KarahNSundsfjordATownerKSamuelsenOInsights into the global molecular epidemiology of carbapenem non-susceptible clones of Acinetobacter baumanniiDrug Resist. Updat.20121523724710.1016/j.drup.2012.06.001
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint tables for interpretation of MICs and Zone diameters. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_8.0_Breakpoint_Tables.xls. (2018).
- HigginsPGDammhaynCHackelMSeifertHGlobal spread of carbapenem-resistant Acinetobacter baumanniiJ. Antimicrob. Chemother.20106523323810.1093/jac/dkp428
- NigroSJHallRMStructure and context of Acinetobacter transposons carrying theoxa23 carbapenemase geneJ. Antimicrob. Chemother.2016711135114710.1093/jac/dkv440
- SiguierPPerochonJLestradeLMahillonJChandlerMISfinder: the reference centre for bacterial insertion sequencesNucleic Acids Res.200634D32D3610.1093/nar/gkj014
- GirlichDDamacenoQSOliveiraACNordmannPOXA-253, a variant of the carbapenem-hydrolyzing class D beta-lactamase OXA-143 in Acinetobacter baumanniiAntimicrob. Agents Chemother.2014582976297810.1128/AAC.02640-133993228
- Dalla-CostaLMOutbreak of carbapenem-resistant Acinetobacter baumannii producing the OXA-23 enzyme in Curitiba, BrazilJ. Clin. Microbiol.2003413403340610.1128/JCM.41.7.3403-3406.2003165295
- MartinsNDalla-CostaLUeharaAARileyLWMoreiraBMEmergence of Acinetobacter baumannii international clone II in Brazil: reflection of a global expansionInfect. Genet. Evol.20132037038010.1016/j.meegid.2013.09.028
- CamargoCHPopulation structure analysis of carbapenem-resistant Acinetobacter baumannii clinical isolates from Brazil reveals predominance of clonal complexes 1, 15, and 79Antimicrob. Agents Chemother.2016602545254710.1128/AAC.02186-154808177
- CardosoJPCayoRGirardelloRGalesACDiversity of mechanisms conferring resistance to beta-lactams among OXA-23-producing Acinetobacter baumannii clonesDiagn. Microbiol. Infect. Dis.201685909710.1016/j.diagmicrobio.2016.01.018
- Coelho-SouzaTLongitudinal surveillance for meningitis by Acinetobacter in a large urban setting in BrazilClin. Microbiol. Infect.201319E241E24410.1111/1469-0691.121453625502
- CorreaADistinct genetic diversity of carbapenem-resistant Acinetobacter baumannii from Colombian hospitalsMicrob. Drug. Resist.201824485410.1089/mdr.2016.0190
- GrossoFOXA-23-producing Acinetobacter baumannii: a new hotspot of diversity in Rio de Janeiro?J. Antimicrob. Chemother.201166626510.1093/jac/dkq406
- RodriguezCHMolecular epidemiology of carbapenem-resistant Acinetobacter baumannii in South AmericaJ. Med. Microbiol.2016651088109110.1099/jmm.0.000328
- StietzMSAcinetobacter baumannii extensively drug resistant lineages in Buenos Aires hospitals differ from the international clones I–IIIInfect. Genet. Evol.20131429430110.1016/j.meegid.2012.12.020
- VasconcelosATThe changing epidemiology of Acinetobacter spp. producing OXA carbapenemases causing bloodstream infections in Brazil: a BrasNet reportDiagn. Microbiol. Infect. Dis.20158338238510.1016/j.diagmicrobio.2015.08.006
- de Sa CavalcantiFLEmergence of extensively drug-resistant OXA-72-producing Acinetobacter baumannii in Recife, Brazil: risk of clonal dissemination?Diagn. Microbiol. Infect. Dis.20137725025110.1016/j.diagmicrobio.2013.07.022
- OpazoAFirst report of blaOXA-23 in Acinetobacter baumannii isolates from Chilean hospitalsJ. Glob. Antimicrob. Resist.20153545510.1016/j.jgar.2015.01.002
- SaavedraSYCayoRGalesACLealALSaavedraCHEarly dissemination of OXA-72-producing Acinetobacter baumannii strain in Colombia: a case reportBraz. J. Infect. Dis.20141867868010.1016/j.bjid.2014.05.017
- PaganoMRochaLSampaioJLMartinsAFBarthALEmergence of OXA-72-producing Acinetobacter baumannii belonging to high-risk clones (CC15 and CC79) in different Brazilian statesInfect. Control. Hosp. Epidemiol.20173825225410.1017/ice.2016.287
- Nunez QuezadaTOutbreak of blaOXA-72-producing Acinetobacter baumannii in South AmericaJ. Chemother.20172932132410.1080/1120009X.2016.1158936
- AlyMGenetic diversity of OXA-51-like genes among multidrug-resistant Acinetobacter baumannii in Riyadh, Saudi ArabiaEur. J. Clin. Microbiol. Infect. Dis.2014331223122810.1007/s10096-014-2068-0
- RafeiRMolecular analysis of Acinetobacter baumannii strains isolated in Lebanon using four different typing methodsPLoS ONE20149e11596910.1371/journal.pone.01159694277430
- de Sa Cavalcanti, F. L. et al. High frequency of OXA-253-producing Acinetobacter baumannii in different hospitals in Recife, Brazil. Antimicrob. Agents Chemother. 61, 10.1128/aac.01309-16 (2017).
- ZanderEBonninRASeifertHHigginsPGCharacterization of blaOXA-143 variants in Acinetobacter baumannii and Acinetobacter pittiiAntimicrob. Agents Chemother.2014582704270810.1128/AAC.02618-133993222
- Mari-AlmirallMMALDI-TOF/MS identification of species from the Acinetobacter baumannii (Ab) group revisited: inclusion of the novel A. seifertii and A. dijkshoorniae speciesClin. Microbiol. Infect.201723210 e211210 e21910.1016/j.cmi.2016.11.020
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Seven Informational Supplement, M100-S28 (Clinical and Laboratory Standards Institute, Wayne, PA, 2018).
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). Recommendations for MIC Determination of Colistin (polymyxin E) as Recommended by the Joint CLSI-EUCAST Polymyxin Breakpoints Working Group. EUCAST.org 1–1 (2016).
- RobledoIEDetection of KPC in Acinetobacter spp. in Puerto RicoAntimicrob. Agents Chemother.2010541354135710.1128/AAC.00899-09
- PoirelLWalshTRCuvillierVNordmannPMultiplex PCR for detection of acquired carbapenemase genesDiagn. Microbiol. Infect. Dis.20117011912310.1016/j.diagmicrobio.2010.12.002
- HigginsPGLehmannMSeifertHInclusion of OXA-143 primers in a multiplex polymerase chain reaction (PCR) for genes encoding prevalent OXA carbapenemases in Acinetobacter sppInt. J. Antimicrob. Agents20103530510.1016/j.ijantimicag.2009.10.014
- HigginsPGOXA-235, a novel class D beta-lactamase involved in resistance to carbapenems in Acinetobacter baumanniiAntimicrob. Agents Chemother.2013572121212610.1128/AAC.02413-123632948
- WoodfordNMultiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter sppInt. J. Antimicrob. Agents20062735135310.1016/j.ijantimicag.2006.01.004
- AltschulSFGishWMillerWMyersEWLipmanDJBasic local alignment search toolJ. Mol. Biol.199021540341010.1016/S0022-2836(05)80360-2
- DurmazRThe optimization of a rapid pulsed-field gel electrophoresis protocol for the typing of Acinetobacter baumannii, Escherichia coli and Klebsiella sppJpn. J. Infect. Dis.200962372377
- DiancourtLPassetVNemecADijkshoornLBrisseSThe population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic poolPLoS ONE20105e1003410.1371/journal.pone.00100342850921
