Abstract
For the first time, we identified 15 cases of Candida auris in Shenyang, China, and then performed a risk factor assessment for these patients compared with 30 control subjects who were hospitalized in the same ward during the same period of time as the infected patients. We found that diarrhea, gastrointestinal decompression, infection, or colonization with other Candida isolates (especially Candida albicans) and tetracycline antibiotics were all risk factors for C. auris infection or colonization. Diarrhea and tetracycline antibiotics were independent risk factors. We suggest clinicians pay special attention to the emergence of multidrug-resistant C. auris infections or colonization.
Introduction
We read with interest the review by Chowdhary and colleagues concerning the rapidly emerging multidrug-resistant pathogenic yeast Candida aurisCitation1, which primarily affects critically ill patients and results in significant morbidity and mortality. We noted that C. auris was first isolated in Japan and described as a new species in 2009Citation2. In 2011, it was identified as a cause of fungemia in South KoreaCitation3. C. auris has caused serious infections globally, including in India, South Africa, Kuwait, the United Kingdom, Venezuela, the United States, Israel, Oman, and, more recently, Panama and the United Arab EmiratesCitation4–Citation13. The real prevalence of C. auris may be underestimated because this pathogen has been routinely misidentified as Candida haemulonii. We therefore reviewed all isolates of Vitek-identified C. haemulonii obtained from a tertiary care hospital in Shenyang, China, and have found 15 misidentified cases. In the present study, we report the first cases of C. auris from China. Moreover, we described in detail the clinical characteristics of patients with a C. auris infection or colonization. Furthermore, to define the risk factors for C. auris infection or colonization, we conducted a matched (1:2) retrospective cohort study using patients who were neither infected nor colonized by C. auris as controls.
Results
Microbiology
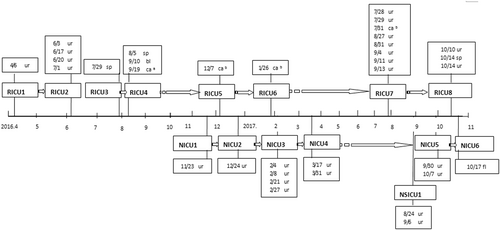
During January 2011 October 2017, 35 isolates were identified as C. haemulonii but later confirmed as C. auris. A total of 26 isolates (74%) were from urine, four from a catheter, three from sputum, one from blood, and one from fluid. These isolates originated from fifteen patients (See Fig. ). For each patient, the first isolate was selected for a follow-up study (apart from RICU4 with blood culture isolates selected).
Onset chart of 15 cases of Candida auris infection (the detection time and sample type are shown in the textboxes); Respiratory ICU inpatient number (RICU1-RICU8); Neurology ICU inpatient number (NICU1–NICU6); Neurosurgical ICU inpatient number (NSICU1); sp sputum; ur urine; bl blood; caa central venous line; cab urinary catheter; dr drainage
Colonies of all isolates on Sabouraud dextrose agar were white to cream colored and smooth. They were able to grow at both 37 °C and 42 °C and had a pink colony color on CHROMagar Candida medium. We observed that organisms from all samples were oval, without pseudohyphae and germ tube formation. All samples were reidentified as C. haemulonii by Vitek-2 (bioMérieux, Marcy I’Etoile, France) and Candida famata by API 20C (bioMérieux) assays. All organisms from our isolate collections could assimilate N-acetyl-glucosamine in contrast to the isolates from Japan and South Korea, which was similar to Chowdhary’s observations in India reported in 2013Citation3. All Vitek-identified C. haemulonii isolates were confirmed to be C. auris by the sequences of the internal transcribed spacer (ITS) region and the D1/D2 region of the large subunit (28S) of the ribosomal DNA. Then, using the genetically identified strains, an in-house super-spectrum for C. auris was created in the RUO (research-use-only) module in the VITEK-MS MALDI-TOF system. MALDI-TOF then could be subsequently used routinely in the laboratory for the rapid identification of C. auris isolates.
Molecular characterization
All of the 15 C. auris isolates had 100% similarity in their ITS, D1/D2, RPB1, and RPB2 sequences. The ITS regions of our C. auris isolates had 100% similarity with isolates from India (GenBank accession KC692039), South Africa (GenBank accession KJ126758/ KJ126761), and Kuwait (GenBank accession LN624638) but 98% sequence homology to isolates from Japan/Korea (GenBank accession AB375772/JX459679). The D1/D2, RPB1, and RPB2 sequences of our isolates had 100% sequence homology to isolates from Japan/Korea (GenBank accession AB375773/ EU881954).
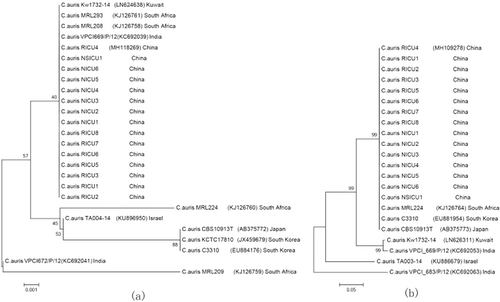
Phylogenetic trees based on the ITS and D1/D2 sequences showed that the C. auris isolates from China were similar to the South African clone, whereas the strain from China was distinct to some extent from other isolates from Israel, India, Japan, and South Korea (See Fig. ).
Phylogenetic trees were generated from the internal transcribed spacer (a) region and D1/D2 domain of the ribosomal DNA large subunit sequences (b). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to each branch. Bold indicates strains from China. GenBank accession numbers are provided in parentheses. Scale bar indicates nucleotide substitutions per site
Antifungal susceptibility testing
All 15 isolates were resistant to fluconazole and susceptible to 5-fluorocytosine, itraconazole (IZ), voriconazole (VOR) and amphotericin B. Notably, all isolates showed high minimum inhibitory concentrations (MICs) to VOR ranging from 0.5 to 1 μg/ml. (Table )
Information and susceptibility test of 15 isolates from 15 patients, Shenyang, China
The first C. auris case report
In order to trace the origin of the first C. auris isolate in China, the complete case progress record was reported here in detail. The first case (See Fig. ) was a 70-year-old man with a 40-year history of high paraplegia and prolonged bed rest who presented with severe pneumonia, respiratory failure, and diabetes complications. He was admitted to the Respiratory and Intensive Care Unit and then treated with empirical broad-spectrum antibiotics, a tracheotomy, an indwelling gastric tube, and urinary catheterization (the urinary catheter was changed once a month). In the hospital on day 1, a urine culture yielded Candida albicans (an azole-susceptible strain), which persisted on days 14, 19, 23, 35, 42, and 45. Antifungal therapy was initiated on day 21 with VOR (400 mg/day) for 7 days. Thereafter, several sputum cultures showed Candida tropicalis and Candida glabrate. On day 47, a blood culture yielded C. albicans (an azole-susceptible strain). On day 50, the antifungal therapy was changed to fluconazole (400 mg/day) for 12 days. During fluconazole therapy, a urine culture again yielded C. albicans (an azole-resistant strain) on day 49. On day 72, the antifungal treatment was changed to caspofungin (50 mg/day) for 10 days. The urinary catheter was removed on day 81, and a culture of the urinary catheter tip removed from the patient grew > 1000 colonies of C. albicans (an azole-resistant strain). On day 88 after admission, a blood culture yielded C. glabrate. On day 92, the antifungal therapy was changed to micafungin (100 mg/day) for 14 days. On day 116, the patient presented with antibiotic-associated diarrhea. On 6 April the 118th day of hospitalization and the 7th day after replacement of the urinary catheter, a urine culture yielded C. auris (fluconazole with MIC > 64 μg/ml). On day 124, the patient was transferred back to a local hospital for treatment.
This patient was treated with voriconazole (VOR 400 mg/day) for 7 days; fluconazole (FLU 400 mg/day) for 12 days; caspofungin (CAS 50 mg/day) for 10 days; and micafungin (MF 100 mg/day) for 14 days. Sp sputum; ur urine; bl blood; ca urinary catheter

Clinical characteristics of C. auris cases
All 15 isolates were recovered from the Intensive Care Unit. Interestingly, all the cases were sporadic, but they appeared consecutively. With the exception of one case from the Neurosurgical ICU (NSICU1), 14 cases of C. auris infection, or colonization were inpatients in either the Respiratory and Intensive Care Unit (RICU1–RICU8) or Department of Neurology and Intensive Care Unit (NICU1–NICU6) (see Fig. ). There was no overlap of the detection time of an infection or colonization except for the RICU7 and NSICU1 samples that came from different departments. There was a 6-month gap between the identification of the RICU6 and RICU7 cases. Cases RICU6 and RICU1 both utilized the same bed during their hospital stay (Bed No. 3), and the other two pairs of cases also used the same bed: RICU2 and RICU8 used Bed No. 7, and NICU2 and NICU6 used Bed No. 12.
All 15 patients had an indwelling urinary catheter (changed once a month), and the average duration of the indwelling urinary catheter was 35 days (range, 6~118 days). All 15 patients had an indwelling gastric tube, 14 patients utilized mechanical ventilation, 10 patients had an endotracheal tube, and 7 patients had a tracheotomy. In total, 10 of the patients had culture-positive urine at least once, and 7 patients (RICU1, RICU2, RICU7, NICU3, NICU4, NICU5, and NSICU1) developed urinary tract infections. A total of three patients (RICU8, NICU1, and NICU2) were considered possibility positive for C. auris colonization because of culture-positive urine samples. In addition, RICU4 was a lung cancer patient who presented with C. auris fungemia on the 35th day after the isolation of C. auris from the sputum, whereas RICU3 was also a lung cancer patient with culture-positive sputum. Moreover, NICU6 had suffered from a biliary tract infection with C. auris. Ten patients had immunosuppressive conditions, seven had diabetes mellitus, three had malignant tumors, and two had chronic kidney disease (Table ).
Univariate risk factor analysis of Candida auris infection cases vs. controls
Risk factor analysis
Five risk factors were statistically significant in the bivariate analysis: diarrhea, gastrointestinal decompression, the presence of other Candida strains, the presence of C. albicans, and the use of tetracycline antibiotics (i.e., minocycline or tigecycline). This study also found that diarrhea and tetracycline (minocycline or tigecycline) therapy were statistically significant in the multivariate analysis (Table ).
Discussion
As it caused successive epidemic outbreaks of fungemia in intensive care unit (ICU) patients in India, Europe, and the United States, multidrug-resistant C. auris has attracted significant concern. At present, cases of C. auris are being reported for the first time in Shenyang, China. Thus, we sought to analyze the risk factors that predisposed a patient for C. auris infection or colonization. In the present study, we analyzed 15 cases of C. auris infection or colonization and compared them with C. auris noninfected or noncolonized patients. The following factors increased the susceptibility to C. auris infection or colonization: diarrhea, gastrointestinal decompression, the presence of other Candida isolates (especially C. albicans), and the use of tetracycline antibiotics. Diarrhea and the use of tetracyclines were independent risk factors.
Both the univariate and multivariate analysis indicated that diarrhea was a risk factor for C. auris infection or colonization; gastrointestinal decompression was also a risk factor in the univariate analysis. Because the first patient experienced diarrhea before the emergence of C. auris infection or colonization, we inferred that the source of C. auris might be the gut, where colonized Candida species could invade via translocation and cause either localized infection or candidemia, as described by Kullberg et al.Citation14. According to Perlin et al., the gastrointestinal tract is an important reservoir of resistance for Candida species, where they can form a mixed biofilmCitation15. This hypothesis remains to be tested through further investigation into the intestinal microecology. C. auris is thought to migrate from the intestine to the urinary system, where it results in infection or colonization, as evidenced by the occurrence of Candiduria in 10 patients in this study. Another study found that C. auris easily aggregates in the mouse kidney, indicating that such aggregation may be a cause of an invasive infection and a continuous infection of C. auris. Ben-Ami et al. observed distinct yeast cell aggregates in the kidneys of mice with fatal C. auris infections, which suggests that aggregation might be a mode of immune evasion and persistence in the tissueCitation7. In the present study, a urinary system infection or colonization lasted for a maximum of 47 d; therefore, it is necessary to establish a renal model of C. auris infection or colonization in order to pursue further research on the pathogenesis of C. auris.
The use of tetracycline antibiotics (tigecycline in five cases and minocycline via intranasal administration in three cases) was statistically significant in both single-factor and multi-factor analyses, but the specific mechanism remains unknown. It is worth investigating whether changes in C. auris’s ecological niches have brought the fungus into greater contact with susceptible humans. The occurrence of C. auris infections or colonization should be closely monitored during the clinical application of tetracycline antibiotics.
In addition, the first patient had infection or colonization with C. albicans and C. glabrate prior to infection or colonization with C. auris. According to the risk factor analysis of cases and controls, a previous C. albicans infection or colonization was a risk factor of C. auris infection or colonization. We think there are two possible reasons. (1) Infection or colonization with other Candida species will increase the dose of antifungal drug required for adequate treatment, and the resulting selection pressure further increases the probability of C. auris infection or colonization. (2) The capability of C. auris to form a biofilm and other virulence traits are weaker than that of C. albicans and other Candida speciesCitation16, so maybe C. auris can cause infection or colonization by obtaining assistance from the invasiveness and mixed biofilm formed by other Candida species such as C. albicans. In the present study, three cases had a C. albicans infection or colonization before the C. auris infection or colonization occurred, and two cases had C. auris infection or colonization and C. glabrate infection or colonization simultaneously.
Historically, the first patient developed C. albicans infection with an azole-resistant strain at 49 d after admission, which lasted for 32 d. Antifungal drugs such as fluconazole, VOR, and echinocandins were administered accumulatively for 51 d, and at 118 d, C. auris infection was observed. Three potential causes can be identified. (1) Did horizontal drug-resistance gene transmission result in the conversion of drug-sensitive C. albicans to drug-resistant C. albicans followed by the conversion to a multidrug-resistant C. auris strain? We are currently pursuing studies that might answer that question. (2) The occurrence of C. auris infection or colonization is significantly associated with the use of antifungal drugs, and this has been confirmed by other studies. Fluconazole has been available since 1991 and echinocandins since the early 2000s; however, these drugs only recently became accessible in resource-limited settingsCitation17. C. auris was first detected in 1996, but it only became globally prevalent during the past 2 years. There is a correlation between the greater prevalence of C. auris and the time antifungal drug treatment was initiated. In the present study, we observed that patients with C. auris infection or colonization were more likely than the control group to have been treated with VOR, but no significant difference was observed because of the small sample size. Therefore, further validation with a larger sample size is required.
The first patient had undergone long-term bed rest (40 years), which does not support acquisition of C. auris by pathways such as foreign travel. In another study, screening for C. auris infection was performed on high-risk patients prior to ICU admission in C. auris epidemic areas. Only one subject in 2246 was found to be carrying C. aurisCitation7. Therefore, C. auris is not likely derived from the community but is, instead, a hospital-acquired pathogen. In the present study, the sequences of the ITS, D1/D2, RPB1, and RPB2 genes in all 15 samples were exactly identical, so we concluded that C. auris was likely disseminated by the same clone in our hospital. Some studies have reported that C. auris can survive for 28 d in the environmentCitation18; however, the interval of onset between the RICU6 and RICU7 samples was 6 months in the present study, suggesting that C. auris can survive far longer than previously reported in the environment. Further investigation is required.
Shenyang (China) is geographically adjacent to Japan and Korea, but the sequencing results for our isolates were more consistent with the isolates from South Africa. Although the reason remains unclear, we speculate that the South African strains had a higher potential for global transmission. More comprehensive studies of the Shenyang C. auris clone would be useful because molecular epidemiological studies may serve as a useful research model for evaluating the potential evolutionary trajectories of C. auris clones.
The present study has several limitations. First, the number of cases is limited, and this should be taken into consideration when interpreting the results. To date, there are only a few studies on the risk factors of C. auris infection or colonization. This study represents a beneficial supplement to the natural history of C. auris or colonization. Second, we performed a retrospective study with all the inherent problems related to that type of study design. The present study findings should be viewed only as preliminary and hypothesis-generating; they require large-scale validation. Third, subsequent research should adopt a better approach for typing methods such as amplified fragment length polymorphism or whole-genome sequencing to determine the clonality of the strains and their relationships to various global clades. Fourth, future studies that emphasize several molecular mechanisms, including resistance genes (ERG11, ERG3, FKS1, FKS2, and FKS3 genes), efflux and transporters, could provide insight about C. auris resistance in China. Fifth, because there was no awareness of the prevalence of C. auris in the present study, there were not any specific infection control measures for this pathogen. There is a requirement for attention to infection control measures to control the spread of C. auris.
We identified one C. auris sample as C. haemulonii by error; thus, we suspect the prevalence of C. auris may be underestimated via misidentification. Further epidemiological studies of C. auris (especially in the ICU) in China should be conducted. Clinicians and microbiologists are now facing urgent challenges stemming from the misidentification of C. auris and decreased susceptibility. We should all be aware of this emerging multidrug-resistant “fake C. haemulonii”, or C. auris yeast.
Materials and Methods
Laboratory methods
We retrospectively reviewed the microbiology records including all isolates identified as C. haemulonii that were later confirmed as C. auris during January 2011 October 2017. These organisms were reidentified by Vitek-2 (bioMérieux, Marcy I’Etoile, France), by API 20C (bioMérieux) assays and by using matrix-assisted laser desorption/ionization time-of-fight (MALDI-TOF) mass spectrometry (bioMérieux, Marcy ľEtoile, France). Subsequent molecular identification was accomplished by sequencing the ITS and D1/D2 regions. A set of four genetic loci, ITS, D1/D2, RPB1, and RPB2, were selected for a multi-locus phylogenetic analysis based on a previously published report by Cendejas-Bueno et al.Citation19. We aligned the ITS and D1/D2 sequences of C. auris isolates with BioEdit and generated phylogenetic trees with the neighbor-joining method. We tested the phylogeny with the bootstrap method (1000 replicates). Evolutionary analyses were performed in MEGA6. In addition, the susceptibility of the isolates was determined using the Sensititre YeastOne colorimetric microdilution method (Thermo Fisher scientific, Oxoid, USA) in an in vitro assay according to the manufacturer’s instructions. Candida krusei ATCC6258 was used as the control strain. Because the breakpoints for C. auris were not defined, the breakpoints suggested for yeast in CLSI M27-S3 were used to interpret the MICs for this new yeast, and CLSI M27-S4 was followed for other Candida speciesCitation20.
Study design
Infected or colonized patients with Vitek-identified C. haemulonii that was later confirmed as C. auris were defined as the case group. Two control subjects hospitalized at the same time and in the same ward (ICU) were selected. These control patients had no detectable C. haemulonii that was later confirmed as C. auris in clinically significant specimens such as sputum, urine, blood, fluid, wound, or stool samples etc.
Patient information
We retrospectively reviewed medical records of the patients including the selected cases and the control group and recorded patient demographics, hospital unit, co- morbidities, medications, and clinical characteristics. Detailed case report forms, designed specifically for this study, were prepared by the physician author (Su fei Tian) and rechecked by a second author (Chen Rong).
Definitions
A candidaemia episode was defined by the isolation of a Candida strain from one or more blood specimen cultures drawn from a peripheral vein. For patients who had more than one episode of fungemia during the same hospitalization, only data from the first episode was analyzed. Possible C. haemulonii that was later confirmed as C. auris infection was defined as a case with a positive culture from a non-sterile site (i.e., sternal wound, urine, vascular line tip) and clinical signs and symptoms of infection requiring treatment with antifungal agents. Colonization with C. haemulonii that was later confirmed as C. auris was defined as culture-positive sputum, urine, blood, pus, wound, stool samples without clinical signs of Candida infection. Exposure to various risk factors was taken into consideration in the retrospective cohort study only if such exposure occurred prior to the development of C. haemulonii infection or colonization that was later confirmed as C. auris infection or colonization.
Standard criteria were used for the definition of existing co-morbidities (i.e., heart dysfunction, diabetes mellitus, chronic renal insufficiency). Steroid treatment was defined as any use of glucocorticoids (i.e., any dose for any period of time) during the hospital stay. The definition of “diarrhea” is “an intestinal disorder characterized by abnormal frequency and fluidity of fecal evacuations”Citation21. According to the Bristol stool scale, Type 5 (Soft blobs with clear cut edges, passed easily), Type 6 (Fluffy pieces with ragged edges, a mushy stool), and Type 7 (Watery, no solid pieces, entirely liquid) were tending toward diarrhea. Moreover, Gram staining was used as a supportive method to help define diarrhea in this study. Gastrointestinal decompression was used to extract the gas and contents of the gastrointestinal tract through a gastric tube, thus facilitating the operation and recovery after the operation.
Exposure to various antimicrobial or antifungal agents was defined by the use of the given drugs for at least 3 consecutive days prior to the development of C. haemulonii infection or colonization that was later confirmed as a C. auris infection or colonization.
Statistical analysis
The data are expressed as the mean ± standard deviation for continuous variables and as percentages for categorical variables. For continuous variables, Student’s t test or the Mann–Whitney U test was, respectively, used for normally and non-normally distributed variables. Categorical variables were compared by χ2 or Fischer’s exact test. Variables with P < 0.1 in the bivariate analysis were included in a backward stepwise multivariate logistic regression model. All statistical analyses were performed using SPSS 22.0 (SPSS Inc., Chicago, IL).
Nucleotide sequence accession numbers
The nucleotide sequences obtained during this study were deposited in the GenBank nucleotide database under accession numbers MH118269, MH109278, MH124606, and MH124607.
Acknowledgements
This study was financially supported by a research grant (81101290) from the National Natural Science Foundation of China and another research grant (2008225010-10) from the Liaoning Science & Technology projects.
Author contributions
Su Fei Tian, Chen Rong, and Yun Zhuo Chu made substantial contributions to the conception and design of the study, or acquisition of the data, or analysis and interpretation of the data; Fu shun Li, Hua Nian, and Shi Tong Cheng were involved in drafting the manuscript or revising it critically for important intellectual content; and Hong Shang provided final approval of the version to be published.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- ChowdharyASharmaCMeisJFCandida auris: a rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globallyPLoS. Pathog.201713 e100629010.1371/journal.ppat.10062905436850
- SatohKCandida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospitalMicrobiol. Immunol.20095341 4410.1111/j.1348-0421.2008.00083.x
- LeeWGFirst three reported cases of nosocomial fungemia caused by Candida aurisJ. Clin. Microbiol.2011493139314210.1128/JCM.00319-113165631
- ChowdharyANew clonal strain of Candida auris, Delhi, IndiaEmerg. Infect. Dis.2013191670167310.3201/eid1910.1303933810747
- MagoboRECorcoranCSeetharamSGovenderNPCandida auris associated candidemia, South AfricaEmerg. Infect. Dis.2014201250125110.3201/eid2007.1317654073876
- EmaraMCandida auris candidemia in Kuwait, 2014Emerg. Infect. Dis.2015211091109210.3201/eid2106.1502704451886
- SchelenzSFirst hospital outbreak of the globally emerging Candida auris in a European hospitalAntimicrob. Resist. Infect. Control201653510.1186/s13756-016-0132-55069812
- CalvoBFirst report of Candida auris in America: clinical and microbiological aspects of 18 episodes of candidemiaJ. Infect.20167336937410.1016/j.jinf.2016.07.008
- VallabhaneniSInvestigation of the first seven reported Cases of Candida auris, a globally emerging invasive, multidrug-resistant fungus-United States, May 2013-August 2016Am. J. Transplant.20171729629910.1111/ajt.14121
- Ben-AmiRMultidrug-resistant Candida haemulonii and C. auris, Tel Aviv, IsraelEmerg. Infect. Dis.20172319520310.3201/eid2302.1614865324804
- Al-SiyabiTFirst report of Candida auris in Oman: clinical and microbiological description of five candidemia casesJ. Infect.20177537337610.1016/j.jinf.2017.05.016
- AraúzABIsolation of Candida auris from 9 patients in Central America: Importance of accurate diagnosis and susceptibility testingMycoses201861444710.1111/myc.12709
- AlatoomAPersistent candidemia despite appropriate fungal therapy: first case of Candida auris from the United Arab EmiratesInt. J. Infect. Dis.201870363710.1016/j.ijid.2018.02.005
- KullbergBJArendrupMCInvasive candidiasisN. Engl. J. Med.20153731445145610.1056/NEJMra1315399
- PerlinDSRautemaa-RichardsonRAlastruey-IzquierdoAThe global problem of antifungal resistance: prevalence, mechanisms, and managementLancet Infect. Dis.201717e383e39210.1016/S1473-3099(17)30316-X
- LarkinEThe emerging pathogen Candida auris: growth phenotype, virulence factors, activity of antifungals, and effect of SCY-078, a novel glucan synthesis inhibitor, on growth morphology and biofilm formationAntimicrob. Agents Chemother.201761e023961610.1128/AAC.02396-165404565
- LockhartSRSimultaneous emergence of multidrug resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analysesClin. Infect. Dis.20176413414010.1093/cid/ciw691
- WelshRMSurvival, persistence, and isolation of the emerging multidrug-resistant pathogenic yeast Candida auris on a plastic health care surfaceJ. Clin. Microbiol.2017552996300510.1128/JCM.00921-175625385
- Cendejas-BuenoEReclassification of the Candida haemulonii complex as Candida haemulonii (C. haemulonii group I), C. duobushaemulonii sp. nov. (C. haemulonii group II), and C. haemulonii var. vulnera var. nov.: three multiresistant human pathogenic yeastsJ. Clin. Microbiol.2012503641365110.1128/JCM.02248-123486233
- Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts: Fourth Informational Supplement M27-S42012Wayne, PA, USACLSI
- SchillerLRGastro 2013 APDW/WCOG Shanghai working party report: chronic diarrhea: definition, classification, diagnosisJ. Gastroenterol. Hepatol.20142962510.1111/jgh.12392
