Abstract
A large German outbreak in 2011 was caused by a locus of enterocyte effacement (LEE)-negative enterohemorrhagic E. coli (EHEC) strain of the serotype O104:H4. This strain harbors markers that are characteristic of both EHEC and enteroaggregative E. coli (EAEC), including aggregative adhesion fimbriae (AAF) genes. Such rare EHEC/EAEC hybrids are highly pathogenic due to their possession of a combination of genes promoting severe toxicity and aggregative adhesion. We previously identified novel EHEC/EAEC hybrids and observed that one strain exhibited aggregative adherence but had no AAF genes. In this study, a genome sequence analysis showed that this strain belongs to the genoserotype O23:H8, MLST ST26, and harbors a 5.2 Mb chromosome and three plasmids. One plasmid carries some EAEC marker genes, such as aatA and genes with limited protein homology (11–61%) to those encoding the bundle-forming pilus (BFP) of enteropathogenic E. coli. Due to significant protein homology distance to known pili, we designated these as aggregate-forming pili (AFP)-encoding genes and the respective plasmid as pAFP. The afp operon was arranged similarly to the operon of BFP genes but contained an additional gene, afpA2, which is homologous to afpA. The deletion of the afp operon, afpA, or a nearby gene (afpR) encoding an AraC-like regulator, but not afpA2, led to a loss of pilin production, piliation, bacterial autoaggregation, and importantly, a >80% reduction in adhesion and cytotoxicity toward epithelial cells. Gene sets similar to the afp operon were identified in a variety of aatA-positive but AAF-negative intestinal pathogenic E. coli. In summary, we characterized widely distributed and novel fimbriae that are essential for aggregative adherence and cytotoxicity in a LEE-negative Shiga-toxigenic hybrid.
Introduction
A large outbreak in 2011 was caused by an enterohemorrhagic Escherichia coli (EHEC) strain of the rare serotype O104:H4, which led to 53 deaths, 855 cases of life-threatening hemolytic uremic syndrome (HUS), and 2,987 cases of gastroenteritisCitation1,Citation2. The outbreak-causing strain was shown to be a hybrid that harbors genes characteristic of both EHEC and enteroaggregative E. coli (EAEC), and it is therefore referred to as EHEC/EAECCitation3,Citation4. The characterization of such hybrid strains is important because of their high pathogenicity, which is a result of the combined production of Shiga toxin and specific adhesins, the latter of which are encoded by aggregative adherence fimbriae (AAF) genes located on the EAEC virulence plasmid pAACitation3,Citation5,Citation6. Specifically, the pAA of the outbreak strain carries genes encoding type I AAF (AAF/I), the AraC-like regulator AggR, and the antiaggregation protein dispersin (Aap) and its export ABC transporter complex (AatA-AatD), among othersCitation7. Some genes encoding classical Shiga-toxigenic E. coli (STEC)/EHEC virulence determinants such as the intimin-encoding gene eaeA, a marker gene for the locus of enterocyte effacement (LEE), the type III-protein secretion system encoding genes within the LEE, and the hemolysin toxin gene (hlyA) typically coded on plasmid, were however not found in the outbreak strainCitation3,Citation4.
EHEC/EAEC hybrid strains have rarely been reported. Such strains are LEE (eaeA)-negative, and intimate attachment to host cells by these strains is triggered by other factors, such as AAF. In 1998, a Stx2-producing and aatA-positive E. coli O111:H2 strain associated with an outbreak of HUS in France was characterized by Morabito et al.Citation8. In 1999, a stx2- and aggR-positive E. coli O86:HNM strain was isolated from a 3-year-old child with HUS in JapanCitation9. However, whether these strains possessed AAF genes was not reported. Dallman et al. described an HUS case in 2012 that was associated with an E. coli O111:H21 isolate expressing stx2 and carrying pAA with genes encoding AAF/V, dispersin, AggR and the Aat complexCitation10. In addition, several O104:H4 EHEC/EAEC strains that predominantly harbored AAF/I genes, although some possessed AAF/III, were described that differed from the 2011 outbreak strainCitation11–Citation17.
To further assess the significance of EHEC/EAEC strains in human disease, we previously reanalyzed STEC/EHEC strains from the German National Reference Center (NRC) for Salmonella and other Bacterial Enteric Pathogens collected between 2008 and 2012 and identified two strains of further interest. Both strains tested negative for eaeA and hlyA and positive for stx2 and the EAEC marker aatA, as well as showed aggregative adhesion to HEp-2 cellsCitation18,Citation19. One of these strains was isolated in 2010 from a case of bloody diarrhea and was shown to belong to the serovar O59:H−[19], MLST ST 1136, and encoded genes for type IV AAF (AAF/IV)Citation19. The other strain was isolated from a case of diarrhea in 2012 (strain 12-05829) and was observed to be nonmotile with rough LPS and belonged to MLST ST 26. Interestingly, strain 12-05829 exhibited strong aggregative adherence but did not possess any known AAF genesCitation19. This strain also did not contain the AggR regulon, which is important for the control of a number of genes involved in virulence encoded on pAA and chromosomal EAEC pathogenicity islandsCitation19,Citation20. EAEC strains negative for aggR are commonly designated as atypical EAECCitation21,Citation22.
The timely detection of Shiga-toxigenic hybrids is important because of their high pathogenicityCitation6. Whereas recognition of stx and its variants is well established, the determination of adhesion or aggregation factors as observed in LEE-negative E. coli pathovars is still challenging due to their diversityCitation23,Citation24. Therefore, understanding such variants and their pathogenicity is essential for EHEC risk evaluation. In this study, we investigated the genetic basis of aggregative adhesion in the Shiga-toxigenic hybrid strain 12-05829 and assessed the consequences of its knockout. As a result, we identified genes for a novel type of fimbriae, designated aggregate-forming pili (AFP), located on a plasmid harboring marker genes for EAEC. We showed that the afp genes are responsible for bacterial piliation, autoaggregation, adhesion, and cytotoxicity and are present in a variety of intestinal pathogenic E. coli from human infections.
Results
Shiga-toxigenic hybrid strain 12-05829 carries EAEC marker genes but not AAF genes
Although the LEE-negative Shiga-toxigenic hybrid strain 12-05829 shows aggregative adherence to HEp2 cells, no genes encoding AAF/I–V were detected by a PCR-based analysisCitation19. Therefore, we performed whole genome sequencing to identify the genes involved in aggregative adhesion. The major genomic characteristics of this strain are summarized in Table and are shown in comparison to the 2011 EHEC/EAEC O104:H4 outbreak strain.
Major genome characteristics and virulence genes of the Shiga-toxigenic hybrid strain 12-05829 and the outbreak strain O104:H4 from 2011 (NCBI: NC_018658.1)
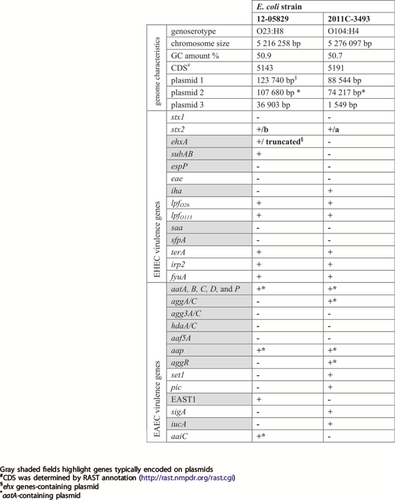
The genome sequence analysis showed that strain 12-05829 belongs to MLST ST26. We also performed genoserotyping by extracting the sequences of the wzx and wzy genes which belonged to serogroup O23. The fliC gene was assigned to serotype H8. Similar to O104:H4, O23:H8 is a very rare serotype, with only one case of human STEC infection with an O23 strain reported in the USA from 2004 until 2014 and only one described by the German NRC from 1997 until 2018Citation25.
Shiga-toxigenic hybrid 12-05829 comprised an ~5.2 Mb chromosome and three plasmids (~124, ~108, and ~37 kb), with the 124 kb plasmid (plasmid 1) exhibiting some features of the hemolysin-coding plasmid of EHEC and the 108 kb plasmid (plasmid 2) harboring aatA (Table , Fig. ). Of note, aatA, which is part of the aatA-D operon located on the pAA plasmid (Fig. ), is commonly used as a diagnostic marker for EAECCitation18.
Plasmid 1 of strain 12-05829 (a) carries genes for the F-pilus, the thin pilus (shown in green) and an incomplete ehx operon (shown in orange) containing a truncated ehxA. Plasmid 2 of EHEC/EAEC strain 12-05829 (b) carries genes typical for EAEC strains (aatA, B, C, D, and P; aap; shown in red) which are also encoded on the pAA plasmid of strain Ec042 (GenBank: NC_017627.1) (c) and additionally contains an operon encoding an aggregate-forming pilus (afp) with limited similarity to the bfp operon of EPEC strains (shown in green) located on the pEAF plasmid (GenBank: NC_011603.1) (d). Plasmid 2 of strain 12-05829, pEAF and pAA all encode AraC-family regulator proteins (shown in blue). e Schematic overview of the afp operon structure of the Shiga-toxigenic hybrid strain 12-05829 compared to the bfp operon of the EPEC strain O127:H6 (80). The afp locus encodes the 14 classical constituents of a pilus locus (afpA - afpL and afpU) and contains an additional gene homologous to afpA, designated afpA2.
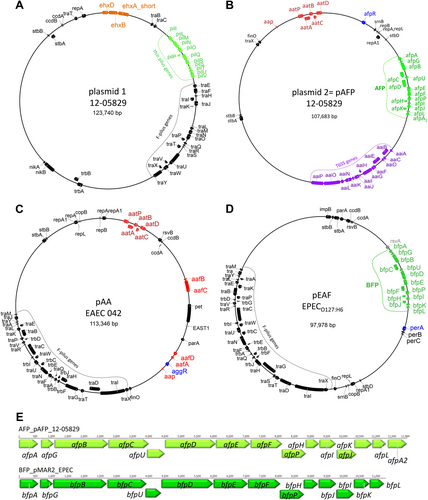
Several chromosomal- or plasmid-located EHEC and EAEC marker genes were identified in strain 12-05829 that were also present in the EHEC/EAEC O104:H4 outbreak strain (Table ). Typical marker genes for EAEC, such as aap (on plasmid 2), and east1 (chromosomal localization) were detected in strain 12-05829 in addition to the aat operon (Table , Fig. ). However, genes coding for the known types AAF (I–V), which are typically located on pAA, were not identified within the aatA-containing plasmid 2 or the chromosome. The structure of the aatA-harboring plasmid 2 of strain 12-05829 compared to pAA of EAEC 042 and pEAF of EPEC O127:H6 is shown in Fig. (Fig. ), as is plasmid 1 of strain 12-05829, which contains parts of the enterohemolysin operon (Fig. ). Of note, plasmid 2 of strain 12-05829 was also observed to carry a putative type VI secretion gene operon (aaiA-P, except aaiM) (Fig. ). Typically, the aai operon is chromosomally localized in EAEC and is implicated in pathogenicityCitation26,Citation27. The aaiC gene encodes a secreted protein and is commonly used as chromosomal EAEC markerCitation27.
Plasmid 1 carries a pil operon that is not essential for aggregative adherence
We subsequently screened the chromosome and plasmid sequences for genes potentially contributing to aggregative adherence. We observed that plasmid 1 of the Shiga-toxigenic hybrid strain 12-05829 carries a type IV pilus biosynthesis locus (pil) consisting of 11 genes (pilL to pilV and pilI) (Fig. ). The pil locus has >99% nucleotide identity to plasmid-coded pil regions of other pathogenic E. coli deposited at NCBI, such as the O113:H2 98NK2 and EH41 STEC strains (GenBank: AF399919 and AY258503). However, deletion of the pil locus in plasmid 1 of strain 12-05829 did not affect bacterial autoaggregation or adhesion to HEp-2 cells (Fig. S1), similar to the results observed for the O113:H2 98NK2 strain after pil plasmid curingCitation28.
Plasmid 2 contains a novel afp operon encoding proteins with limited similarity to the bundle-forming pilus of EPEC as well as a novel AraC-like regulator gene
Interestingly, the aatA-containing plasmid 2 (Fig. ) was observed to carry genes encoding proteins with some homology to the bundle-forming pilus (BFP) apparatus of enteropathogenic E. coli (EPEC). Typical EPEC strains contain an EPEC adherence factor plasmid (pEAF) with a bfp operon coding for type IV pili (Fig. )Citation29. BFP are required for the formation of adherent microcolonies in a pattern known as localized adherence and for full EPEC virulenceCitation29. Fourteen genes (bfpA, G, B, C, U, D, E, F, P, H, I, J, K, and L) are arranged in an operon in pEAF, such as pMAR2 of EPEC O127:H6, as well as three genes coding for regulatory proteins (bfpT, V, and W; designated perA, B, and C in pMAR2) that are necessary for the formation of functional BFPCitation29. A similarly organized operon consisting of 15 genes, designated here as AFP, was identified on plasmid 2 of EHEC/EAEC 12-05829. Therefore, plasmid 2 was named pAFP (Fig. ). Interestingly, two homologues of the EPEC bfpA gene are present in the afp operon at the first (designated afpA) and last position (designated afpA2) (Fig. , Supplementary Table S1). The average nucleotide identity of this novel operon compared to bfp operons described by Nataro et al. in 1987 and Tobe et al. in 1999 was approximately 52% (36-59%)Citation30,Citation31 (Supplementary Tables S1 and S2), whereas the identity between classical bfp operon genes is typically >98% (Supplementary Table S2). The limited relatedness of the classical EPEC bfp genes to the novel afp genes of strain 12-05829 explains why it was previously not possible to detect the afp operon by means of routine PCR based on the bfpA sequence from EPEC strain B171Citation32. In EPEC, bfpA encodes the major structural subunit of the bundle-forming pilusCitation29. Although the protein relatedness was even lower (average 42%, 18–60%) than nucleotide identity, many AFP proteins share conserved domains with BFP proteins. This observation suggests that although AFP are clearly distinct from EPEC BFP, they may have similar functions (Supplementary Table S1).
The expression of the EPEC bfp operon is regulated by BfpT/V/W, which is also designated as PerA/B/C and is encoded on pEAF (Fig. )Citation29. BfpT (PerA) belongs to the AraC family of transcriptional activators and is required for the autoactivation of the per operonCitation29. The aatA-containing plasmid 2 (pAFP) of strain 12-05829 was not observed to harbor an EAEC per operon or aggR but was shown to carry a gene encoding a protein with an AraC family regulatory helix-turn-helix domain (Fig. ). The novel putative AraC-regulator, designated as AfpR, is only 28% identical to BfpT (PerA) of the classical EPEC and 38% identical to AggR of EAEC. Thus, we identified a novel aatA-containing-plasmid-localized afp operon encoding proteins with limited similarity to the bundle-forming pilus of EPEC as well as a novel putative AraC-like regulator gene.
Identification of novel pilus structures that promote a high degree of autoaggregation
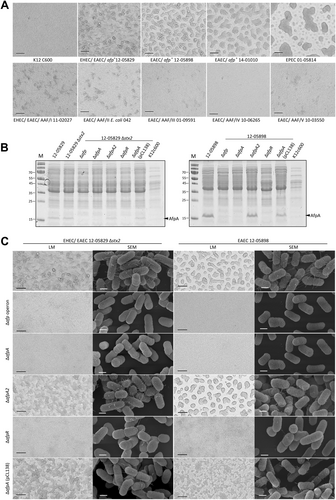
As typical EPEC and EAEC strains are capable of autoaggregationCitation20,Citation23,Citation29,Citation33, we wanted to assess the degree and pattern of autoaggregation for strains containing the afp operon. Specifically, we compared the Shiga-toxigenic hybrid strain 12-05828 and two additional afp-positive E. coli strains harboring aatA but not the AAF genes from the NRC collection (strains 12-05898 and 14-01010) to typical EPEC, EAEC strains with AAF I–V, and a negative control E. coli K12 C600 strain (Table ).
Selected E. coli strains used in this study
We observed a strong degree of autoaggregation for all afp-positive strains after 3 h of incubation but not for the K12 C600 strain. The autoaggregation phenotype of the afp-positive strains (12-05829, 12-05898, and 14-01010) did not resemble that of the AAF I–V-positive EAEC, which produced smaller and more diffuse aggregates, but the phenotype was similar to that of the EPEC strain producing more prominent aggregates (Fig. ). Although this result suggested a functional similarity between AFP and BFP, which was also suggested by their shared protein domains, a clear phenotypic difference was observed between AFP and AAF. Furthermore, we constructed mutants in two afp-positive strains (Shiga-toxigenic hybrid 12-05829 Δstx2 and EAEC 12-05898) in which the entire afp operon, afpA, afpA2, or afpR were deleted. To verify the mutants, the presence of the major pilin AfpA was assessed by SDS-PAGE. We detected the characteristic ~18 kDa protein in all samples except in the ∆afp operon, ∆afpA, and ∆afpR mutants, and complementation was achieved by the introduction of the pCL138 plasmid into the ∆afpA mutant (Fig. ). Accordingly, we observed a lack of autoaggregation in the ∆afp operon, ∆afpA, and ∆afpR strains but not in the ∆afpA2 knockout mutant (Fig. ). Scanning electron microscopy (SEM) observations revealed the presence of fimbriae-like structures on highly aggregating bacteria of both the afp-positive unmodified bacterial strains and the ∆afpA2 mutant, but they were absent in the ∆afp operon and ∆afpA mutants and exhibited a reduced occurrence in the ∆afpR mutant (Fig. , more detailed presentation of pilus in Fig. S2). Reintroduction of the afp operon on plasmid pCL138 led to a restoration of aggregates and fimbriae in the ∆afpA mutants of both strain types (Fig. ). In summary, we showed that afp-positive strains displayed a high degree of autoaggregation that depended on the presence of a functional afp operon, the major pilin-encoding gene afpA and the regulatory protein-encoding gene afpR, but not afpA2.
a Light microscopy images of autoaggregating E. coli K12 C600, the afp-positive strains 12-05829, 12-05898, 14-01010, EPEC 01-05814 and AAF/I–V-positive strains. Formation of large bacterial aggregates is evident for the afp-positive strains 12-05829, 12-05898 and 14-01010 as well as for the EPEC strain 01-05814. b Cell lysate proteins of afp-positive E. coli strains 12-05829 and 12-05898, their respective afp, afpA, afpA2, and afpR deletion mutants and the ΔafpA complementation strain carrying pCL138 separated by SDS-PAGE. c Autoaggregation of the afp-positive E. coli strains 12-05898, 12-05829 Δstx2, the respective afp, afpA, afpA2 and afpR deletion mutants and the ΔafpA complementation strain carrying pCL138. Light microscopy (LM) images for (a) and (c) were captured with 200-fold magnification (scale bar 100 µm) and scanning electron microscopy images for (c) with 50,000-fold magnification (scale bar 500 nm). M molecular weight marker in kDa
Afp-positive E. coli strains exhibit aggregative adherence to epithelial cells that is dependent on the afp operon, afpA, and afpR but not afpA2
We analyzed the pattern and degree of adherence of afp-positive E. coli to HEp-2 cells compared to that of AAF/I–V-positive EAEC, a BFP-positive EPEC strain, and a negative control E. coli K12 C600 strain. Whereas the K12 C600 strain did not show any adherence, the afp-positive strains exhibited a high degree of adherence (Fig. ). The adherence pattern of the afp-positive strains was more similar to that of the EPEC strain than to AAF/I–V-positive EAEC isolates. However, it is important to note that the aggregates of the afp-positive strains (12-05829, 12-05898, and 14-01010) on HEp-2 cells were unusually large and did not completely resemble those observed for EPEC, indicating that AFP indeed represent a distinct type of pili (Fig. ).
a Aggregative adherence to HEp-2 epithelial cells of the afp-positive E. coli strains 12-05829, 12-05898 and 14-01010 compared to EPEC 01-05814 and AAF/I–V-positive strains. Strain K12 C600, which lacks aggregative adherence, served as negative control. b Aggregative adherence to HEp-2 epithelial cells of the afp-positive E. coli strains 12-05898, 12-05829 Δstx2, and the respective afp, afpA, afpA2 and afpR deletion mutants and the ΔafpA complementation strain carrying pCL138. Phase contrast microscopy images are shown with 1000-fold magnification (scale bar 25 µm) for (a–c) Quantitative HEp-2 adhesion assay of the Shiga-toxigenic hybrid strain 12-05829, the respective stx2, afp, afpA, afpA2, afpR deletion mutants and the ΔafpA complementation strain carrying pCL138. Bars show the percent of bacteria adhered to HEp-2 cells after 4 h. d Cytotoxicity assay performed with EHEC/EAEC strain 12-05829, its respective stx2, afp, afpA, afpA2, afpR deletion-mutants and the ΔafpA complementation strain carrying pCL138. The results for (c) and (d) represent the means and standard deviations of triplicate reactions and are representative for at least two additional experiments
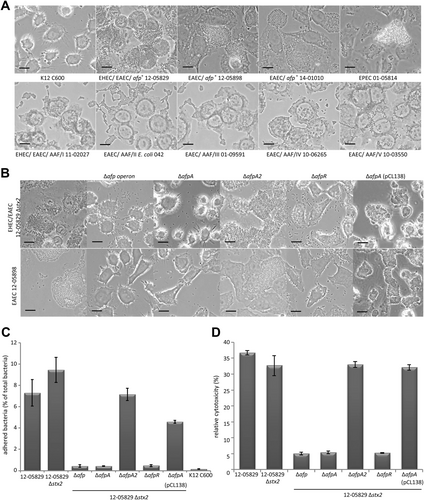
Subsequently, we tested the abovementioned mutants of the Shiga-toxigenic hybrid 12-05829 Δstx2 and EAEC 12-05898 strains for adherence to HEp-2 cells. We observed a lack of adhesion by the strains lacking ∆afpA and the ∆afp operon but not for the ∆afpA2 knockout mutant (Fig. ). In addition, the deletion of afpR led to decreased adhesion. The reintroduction of the afp operon on a plasmid into the ∆afpA mutants restored the adhesion pattern for both strains (Fig. ). To quantify the extent of the adhesion, we performed an assay in which infected HEp-2 cells were washed and the amount of remaining bacteria was determined after 3 h of incubation (Fig. ). This assay clearly demonstrated that the lack of afpA, the afp operon, or afpR but not stx2 or afpA2 led to dramatically reduced adherence of bacteria to HEp-2 cells (Fig. ). The E. coli K12 C600 strain did not show adhesion, and complementation of the ∆afpA mutant using pCL138 reestablished adhesion (Fig. ). In summary, we showed that afp-positive strains exhibited aggregative adhesion to HEp-2 cells and formed unusually large aggregates that depended on the afp operon, the major pilin gene afpA, and afpR but not afpA2.
AFP are essential for cytotoxicity toward epithelial cells
In addition to being important for adhesion to host cells, pili may also cause cytotoxicityCitation34,Citation35. To assess the role of AFP in cytotoxicity, HEp-2 cells were incubated for 24 h with the Shiga-toxigenic hybrid strain 12-05829 and its mutants. The relative cytotoxicity was quantified via the detection of free lactate dehydrogenase from leaky HEp-2 cells. The results demonstrated the cytotoxicity of the wild-type strain, its stx2, and afpA2 negative mutants and the afp complemented ∆afpA mutant, whereas the ∆afpA, ∆afp operon, and ∆afpR mutants were clearly less cytotoxic toward HEp-2 cells (Fig. ). These results demonstrate that AFP are essential for mediating cytotoxicity toward HEp-2 cells.
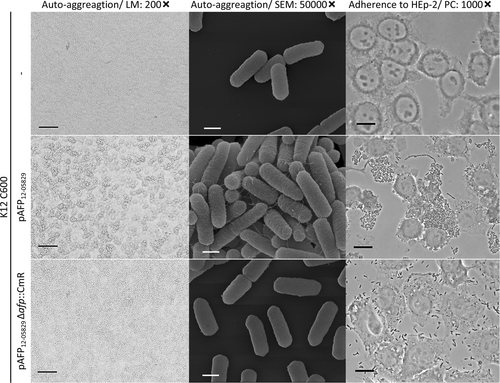
Transformation of pAFP12-05829 into afp-negative E. coli K12 leads to bacterial autoaggregation and aggregative adherence toward epithelial cells
Next, we tested whether the plasmid containing the afp operon and its associated phenotypes, such as autoaggregation and HEp-2 cell adhesion, could be transferred to an afp-negative E. coli strain. Thus, the plasmid pAFP12-05829 was labeled with a CmR cassette and isolated from the host strain for transformation into the afp-negative E. coli K12 C600 strain. Indeed, after the introduction of pAFP12-05829, the K12 C600 strain exhibited characteristic autoaggregation, fimbriae, and aggregative adhesion to HEp-2 cells (Fig. ), whereas the deletion of the afp operon caused loss of these phenotypes (Fig. ). This demonstrated that pAFP and the afp operon it harbors is sufficient to mediate autoaggregation and host cell adherence in other E. coli strains.
The autoaggregation and aggregative adherence to HEp-2 epithelial cells are shown for K12 C600, K12 C600 (pAFP12-05829) and its respective afp deletion plasmid. Light microscopy (LM) images with 200-fold magnification (scale bar 100 µm), scanning electron microscopy (SEM) images with 50,000-fold magnification (scale bar 500 nm) and phase contrast microscopy images (PC) with 1000-fold magnification (scale bar 25 µm) are shown
afp operons are present in a variety of aatA-positive E. coli strains from human infections
We next assessed how widely distributed the afp operon is in the genomes of other E. coli strains. Using an NCBI nucleotide BLAST analysis with the strain 12-05829 afp operon as a query sequence, 17 afp-positive E. coli strains were identified (Supplementary Table S3) from all available E. coli, Shigella and Citrobacter genomes. Compared with strain 12-05829, the nucleotide identity of these afp operons compared was approximately 95.6–98.6%, confirming their close relatedness (Supplementary Table S4). All afp-positive strains were additionally positive for the aat operon, aap, and the gene encoding the new type of AraC family regulator (afpR), but these strains tested negative for stx, eaeA, hlyA, aggR, and bfp (Supplementary Table S3). Furthermore, in the NRC strain collection, we identified 35 aatA-positive but AAF/I–V gene- and stx-negative E. coli strains isolated from human infections between 2001 and 2014 (Table and Supplementary Table S5). To our surprise, 26 of the strains were positive for the afp operon when analyzed for afpB, D, P, A2 and R by PCR (Table and Supplementary Table S5), indicating that E. coli strains harboring afp are regularly associated with human disease. Interestingly, these strains belong to a broad variety of serotypes and MLST STs (Table and Supplementary Table S5). The genome of one of these strains (12-05898), which was used in this study for phenotypic assays (Figs. and ), was sequenced for a more detailed analysis of the aatA-containing pAFP. The MiSeq reads obtained for this strain were mapped to the pAFP12-05829 sequence and covered 90.7% of the reference with a pairwise identity of 95.5%. Importantly, the entire aat operon (nucleotide identity of 98.56%), aap, afpR and the aai operon genes were present in the sequence data of strain 12-05898.
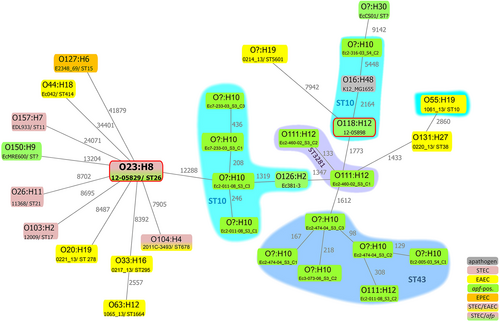
The chromosome sequence of the Shiga-toxigenic hybrid strain 12-05829 was phylogenetically compared to selected EHEC/STEC (O26:H1, O103:H2, and O157:H7), EAEC (O44:H18 strain 042 and recently published EAEC genomesCitation36), STEC/EAEC (O104:H4), EPEC (O127:H6) and other afp-positive E. coli strains (Table and Supplementary Table S4), as well as the nonpathogenic E. coli K12 MG1655 (O16:H48) strain using SNP-based analysis (Fig. ). Although still considerably distant, the most closely related E. coli genome to strain 12-05829 among these strains was that of EHEC/EAEC O104:H4 from 2011. SNP analysis further revealed that the E. coli strains did not cluster due to their pathovar or the presence of afp genes (Fig. ). In summary, the novel afp operon is present in a variety of other aatA-positive E. coli strains, and the Shiga-toxigenic hybrid strain 12-05829 is only distantly related to other strains harboring the afp operon.
Chromosomal phylogeny of afp-positive E. coli strains and examples of EAEC, STEC, EPEC and EHEC/EAEC strains in relation to the afp-positive EHEC/EAEC strain 12-05829 are shown, represented as a minimum spanning tree based on 94 553 core SNPs of 31 strains. The different E. coli strains are phylogenetically diverse or cluster due to their sero- or MLST sequence-type, but not due to their pathovar. All sequences, except those of strain 12-05829 and 12-05898 (red frame), were obtained from NCBI (Supplementary Table S3). The gray numbers next to the lines represent the SNP count. ST? = unassigned MLST ST. O? unassigned serogroup
Discussion
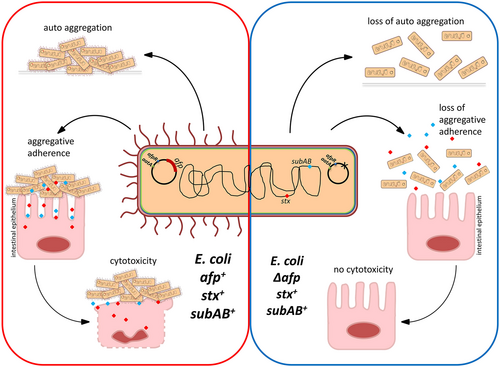
In this study, a detailed genomic analysis of the Shiga-toxigenic hybrid strain 12-05829 resulted in the identification of an aatA-containing plasmid, pAFP, encoding the novel pili locus afp, which is essential for bacterial aggregation, aggregative adherence to host cells, and mediation of cytotoxicity (Fig. ). Interestingly, afp-encoded proteins showed a high degree of identity (average of ~99%) among different afp-positive strains but a lower identity (average of ~42%) to EPEC BFP proteins (Supplementary Table S2), indicating that the novel AFP are only distantly related to BFP.
Red and blue dots represent potential toxins, such as SubAB or Shiga toxin
The afp locus encodes the 14 classical constituents of a pilus operon (afpA to afpL and afpU) as well as an additional homologue of the major pilin gene afpA, afpA2 (Fig. )Citation29. Although AfpA and AfpA2 share ~60% protein identity, AfpA was the only abundant major pilin detected in this study that contributed to AFP pilus generation and the consequent autoaggregation, adhesion, and cytotoxicity phenotypes. Therefore, the role of AfpA2 is still unclear and remains a subject for further research.
AFP pili exhibited considerable cytotoxicity toward HEp-2 cells (Fig. ). On the one hand, pili promote clustering of bacterial cells into large aggregates that nearly cover the entire host cell, allowing bacteria to secrete toxins at high localized concentrations. In addition, the clusters further hamper the diffusion of toxins to the cell exterior. On the other hand, pili assure close proximity between bacteria and the host cell, which may allow the efficient application of secreted toxins and other factors directed toward the host cell (Fig. ). Similar effects to those that were observed for AFP have been described for EPEC. An EAF-negative EPEC strain not expressing BFP was observed to be less cytotoxic than EAF-positive EPECCitation35. Pili-mediated cytotoxicity is not limited to intestinal pathogenic E. coli. Recently, Basso et al. showed that pili promote close contact between bacteria and eukaryotic cells and the localized action of the exolysin ExlA during infection with Pseudomonas aeruginosa. Such an intimate interaction is required for the formation of pores and subsequent eukaryotic cell deathCitation37, a scenario that may be of similar importance for the Shiga-toxigenic hybrid described in this study.
In our experiments, no major difference in cytotoxicity between the afp-positive Shiga-toxigenic strain 12-05829 and its stx2 knockout mutant was noted (Fig. ). This observation may be explained by the far lower toxicity of Stx2b of strain 12-05829 compared to Stx2aCitation38, which is caused by the weak binding of Stx2b to the Shiga toxin receptor globotriaosylceramide Gb(3)Citation39 and its limited capability for proteolytic activation by furinCitation38. These observations raise the question of what other toxin may be responsible for the observed cytotoxicity? The Shiga-toxigenic hybrid strain 12-05829 possesses subAB (Table ), encoding the subtilase AB5 cytotoxin produced by some STEC, especially LEE-deficient strainsCitation40. Because SubAB is known to induce apoptosis and cell death in HeLa cellsCitation41, we hypothesize that SubAB has a toxic effect on HEp-2 cells triggered by AFP. Another factor that may be involved in the observed cytotoxicity is EAST1 (EAEC heat stable enterotoxin), encoded by astA, which activates membrane-bound guanylate cyclase and causes downstream effects on several signaling pathways, ultimately leading to loss of electrolytes and water in intestinal cellsCitation42.
We also identified and characterized a gene encoding a putative regulator of the afp operon in pAFP, designated afpR, which similar to AggR from EAEC and BfpT from EPEC contains an AraC-regulator family conserved domain (Fig. )Citation43. Indeed, pilus structures and pilin protein, as well as the associated autoaggregation, adhesion, and cytotoxicity were reduced in the ∆afpR mutants of afp-positive E. coli strains. The effects of the afpR gene deletion confirm that the novel AraC regulator family protein is important for the formation of pili and likely plays a similar role as BfpT (PerA) in EPEC and AggR in EAECCitation44,Citation45. The presence of a gene that is homologous to afpR on a plasmid with aatA and aap may therefore be of relevance in atypical EAECCitation21,Citation22. Atypical EAEC strains are characterized by a variety of EAEC marker genes, such as aatA and aaiC but are negative for aggR. It is therefore conceivable that another AraC family regulator, such as the one identified in this study, may regulate gene expression. This possibility can now be easily tested using the PCR method described in this study (Supplementary Table S6). Atypical EAEC are frequently identified, and the aatA/afp-positive pathogenic E. coli described here likely belong to this category. For example, in a Brazilian study of the etiology of acute diarrhea in children, 8% of the aatA-positive EAEC strains were aggR-negativeCitation26. In a study by Lima et al. 2013, the classical aggR gene was only observed in 56.7% of the EAEC-positive samples from BrazilCitation27, raising the question of whether the residual >40% of samples contained afpR. Further investigations of the AfpR regulator are currently in progress to examine whether it acts similar to AggR, which activates the expression of many genes in EAEC strain 042, including chromosomal and plasmid-borne lociCitation44,Citation46,Citation47.
The afp locus was also identified in 17 strains deposited at NCBI and in 26 aatA-positive and AAF/I–V gene-negative E. coli from the German NRC strain collection. Of these 43 strains, all were aatA-positive, suggesting that the afp operon is located on a similar pAFP plasmid (Table , Supplementary Tables S3 and S5). Indeed, an alike pAFP was detected in the EAEC strain 12-05898 investigated in this study, because the complete afp operon was present and the MiSeq reads that covered more than 90% of the pAFP12-05829 sequence. This result is in accordance to the observation that afp-associated properties may be transmitted by means of plasmid transfer to other E. coli (Fig. ). For the remaining 25 afp- and aatA-positive strains from the NRC collection we confirmed afp operon plasmid localization, because the presence of afpB, afpD, afpP and afpA2 were detected by PCR with isolated plasmid DNA. A phylogenetic comparison of the afp-positive strains did not reveal a close relationship (Fig. ). Therefore, it is conceivable that the afp-containing plasmid is taken up by E. coli strains, such as by conjugative plasmid transfer. In summary, our findings demonstrate that the novel afp locus is widely distributed among aatA-positive and AAF/I–V gene-negative E. coli strains and is present in a novel plasmid among aatA-containing plasmids that are unrelated to the aatA- and AAF-containing plasmid pAA of EAEC and the aatA-negative bfp-containing plasmid pEAF of EPECCitation48,Citation49.
None of the aatA- and afp-positive strains from NCBI or the NRC collection were stx-positive, indicating that the Shiga-toxigenic hybrid strain 12-05829, such as the outbreak strain from 2011, represents the rare occurrence of hybrid strains combining stx2 with an extraordinarily strong aggregative adherence. The clinical symptoms of the patient carrying strain 12-05829 at the time of sampling were diarrhea without the excretion of blood or HUS development. Strain 12-05829 carries the stx2b gene, and the stx2b-derived toxin is known to be at least 25-times less potent than those derived from stx2a and stx2dCitation38. Nevertheless, stx2b-containing strains have been isolated from HUS patients at NRC in two cases, one strain also represented in the HUSEC collection (HUSEC028) harboring stx2b in combination with stx1c and another previously reported strain harboring stx2b in combination with stx1aCitation11,Citation38,Citation50,Citation51. However, since the afp operon-carrying strains appear diverse, other types of stx with more severe pathogenic potential may also be acquired by means of bacteriophage incorporation. In contrast, STEC strains may also take up pAFP to generate STEC/AFP hybrids, which may have a higher virulence potential.
In summary, in this study we showed that AFP are important and widely distributed adhesion and virulence factors that should be included into the adhesion factor panel used to analyze LEE-negative STEC and for risk evaluation. As the AFP genes studied here are located on a novel aatA-containing plasmid, horizontal gene transfer of the AFP operon to other E. coli strains and even other species is conceivable and may be important for aggregative adherence phenotypes, as shown for the E. coli K12 C600 strain (Fig. ) and to increase toxic effects.
Materials and Methods
Strains used in the study
The strains used in the study are listed in Supplementary Table , Supplementary Tables S5 and S7. The strains outlined in Table and Supplementary Table S5 were, in most cases, collected by the German National Reference Centre (NRC) for Salmonella and other Bacterial Enteric Pathogens and were routinely analyzed for the phenotypic serovar, stx gene type, eaeA, AAF type and MLST ST as described previouslyCitation19. Unless otherwise stated, strains were grown on nutrient agar (Oxoid GmbH, Germany) or Luria Bertani (LB) broth or agar.
Whole-genome sequencing (WGS)
Whole-genome sequencing of Shiga-toxigenic hybrid strain 12-05829 was performed by GATC Biotech (Konstanz, Germany) using a PacBio RS II sequencer (Pacific Biosciences, USA), which produces long read sequences. DNA was isolated by using a Qiagen Genomic-Tip 100/G Kit (Qiagen, Germany) according to manufacturer’s instructions, and 10 µg was sent to GATC. Additionally, short read genome sequencing was performed using an Illumina MiSeq benchtop sequencer in paired-end mode with a v3 chemistry-based cartridge 600 (600-Cycle Reagent Kit, Illumina, Germany). In this case, DNA from the E. coli strains EHEC/EAEC 12-05829 and EAEC 12-05898 was isolated with a Qiagen DNeasy Blood & Tissue Kit (Qiagen, Germany) according to manufacturer’s instructions, and 1 ng of the extracted DNA was used to generate libraries using the Nextera XT DNA Library Preparation Kit from Illumina (FC-131-1024). The sequences were uploaded to the European Nucleotide Archive (ENA) in study Acc. No. PRJEB28343 (Acc. no. ERS2673049 for MiSeq reads of strain 12-05829; Acc.no. ERS2673050 for MiSeq reads of strain 12-05898; and Acc.no. ERS2673049 for PacBio contigs of strain 12-05829).
Bioinformatics analyses
De novo assembly of the PacBio sequence data was performed by GATC utilizing HGAP3 (Pacific Biosciences). The polished assembly yielded 3 plasmid contigs and 5 contigs belonging to the chromosome with a 41- to 63-fold coverage. Based on the overlapping ends of the 3 plasmid contigs, it was possible to close the plasmid sequences, which was verified by PCR (for primers see Supplementary Table S6).
EHEC, EAEC, and enteropathogenic Escherichia coli (EPEC) marker genes (Table ) were searched for by mapping the trimmed Illumina reads against the respective gene sequences downloaded from NCBI with Geneious R10.0.5 (Biomatters Limited, Auckland, New Zealand) (map to reference function), where a coverage of 100% of the reference sequence and 95% sequence homology was set as the threshold. Second, we checked for homologs of the AAF/I to AAF/V and BFP genes using Geneious R10.0.5 (annotate function) with a cut-off of 80% identity. Third, we translated the open reading frames of the Shiga-toxigenic hybrid strain 12-05829 plasmids 1 and 2 into protein sequences and searched for homologs using NCBI pBLAST (standard settings). Using the third strategy, the pil genes were identified on plasmid 1, and the novel afp genes with limited homology to bfp were identified on plasmid 2. An in-house pipeline was used for phylogenetic analysis, including (I) read trimming using Trimmomatic (vers. 0.32) with default parameters, (II) alignment of trimmed reads to reference sequence using BWA mem with default parameters (version 0.7.10-r789), (III) sam file to bam file conversion using samtools (version 0.7.10-r789), (IV) pileup using samtools mpileup (without probabilistic realignment for the computation of base alignment quality), (V) variant calling using VarScan (vers. 2.3: parameters: min-coverage, 10; min-reads, 8; min-avg-qual, 20; min-var-freq, 0.8; min_freq-for-hom, 0.8; p-value, 0.01; and strand-filter disabled), and (VI) consensus sequence creation. To build the consensus sequences, insertions were excluded, and base calls were only considered if supported by at least 80% of the reads (otherwise N was called). SNPs were filtered using a previously published SNP filterCitation52.
The pseudosequences of polymorphic positions were used to create a minimum spanning tree with PhylovizCitation53. To include complete genome sequences from NCBI to phylogenetic analysis, the sequences were converted to artificial FASTQ reads using artfastqgeneratorCitation54 and were mapped to the reference sequence without quality-based trimming.
Nucleotide and protein sequences were compared using MAFFT alignment within Geneious R10.0.5.
Construction of genetically modified strains
Deletion mutants (Supplementary Table S7) were constructed according to the method described by Datsenko and WannerCitation55 using the primer sets listed in Supplementary Table S6 to amplify a FRT-flanked-CmR-gene with homology extensions to the respective gene region to be deleted. After selecting the CmR-positive deletion mutants, the CmR gene was eliminated using the helper plasmid pCP20Citation55. After the CmR-gene was eliminated, the strains were tested for the correct gene deletion by Sanger sequencing using the primer sets indicated in Supplementary Table S6. To transfer the pAFP12-05829 plasmid into a recipient strain, a transposase located on the plasmid was replaced by a CmR gene as mentioned above. The pAFP12-05829 Δtransposase::CmR plasmid was purified and introduced into the K12 C600 recipient strain by electroporation with a Life Technologies Cell-Porator according to the manufacturer’s specifications as previously describedCitation56. After elimination of the CmR gene, the afp deletion was introduced, resulting in K12 C600 (pAFP12-05829 Δtransposase Δafp::CmR). To complement the afpA deletion, the afp operon of strain 12-05829 was cloned into pBeloBac11 via Gibson Assembly using the primer set indicated in Supplementary Table S6Citation57.
PCR for the detection of afpB, afpD, afpP, afpA2 and afpR
PCR was performed using primer sets shown in Supplementary Table S6. Each reaction contained 2.5 μl of 10 × PCR buffer (NEB, Germany), 1 unit of Taq DNA polymerase (NEB, Germany), 5 pmol of each forward and reverse primer, 200 μM of each deoxynucleoside triphosphate (Bioline, Germany) and distilled water to a total reaction volume of 15 μl. A small amount of a single bacterial colony resuspended in 10 µl of distilled water and heated for 10 min at 95 °C or 10 ng plasmid DNA was used as DNA template. DNA amplification was performed in a PCR thermal cycler using the following conditions: 94 °C for 5 min, followed by 30 cycles of 30 seconds at 94 °C, 1 min at 55 °C, and 1 min at 72 °C, with a final extension of 5 min at 72 °C.
SDS-PAGE
Ten microliters of an overnight culture of E. coli was incubated in 1 ml of DMEM (with 0.45% glucose, Lonza, Germany) containing 1% mannose at 37 °C and 5% CO2 for 4 h. Subsequently, the cells were pelleted, lysed in 75 µl of BugBuster (Novagen, Germany) plus 25 µl of 4× Laemmli Buffer, and heated to 95 °C for 10 min. Fifteen microliters of each sample was loaded onto a 15% SDS-polyacrylamide gel, and Coomassie staining was performed after electrophoresisCitation58.
Bacterial autoaggregation assay
For the autoaggregation assay, bacteria were grown overnight in LB and then were inoculated 1:100 into 1 ml of DMEM (with 0.45% glucose, Lonza, Germany) containing 1% mannose in a 24-well plate and incubated for 3 h statically at 37 °C. Light microscopy was performed to detect the autoaggregation phenotype using a Nikon Eclipse inverted microscope (Nikon Instruments, Germany) at 200-fold magnification. For higher resolution, aggregates were analyzed by scanning electron microscopy (SEM). Round glass coverslips (12 mm, Thomas Scientific, Germany) were placed into the wells of a 24-well plate and autoaggregation was assessed as described above. K12 C600 bacterial cells were applied to plastic culture dishes (IBIDI dish; µ-Dish 35 mm, high, IBIDI, Germany) for aggregation assay in a volume of 3 ml. At the end of the aggregation assay, bacteria were fixed in 1% paraformaldehyde, 2.5% glutardialdehyde, 0.05 M HEPES buffer (pH 7.2) for 2 h at room temperature and then were gently washed with distilled water prior to postfixation in 1% OsO4 (1 h). Samples were again washed in distilled water, dehydrated in an ethanol series (30, 50, 70, 90 and 96% for 15 min each and absolute ethanol for 30 min) and then were critical point dried (CPD 300, Leica, Germany) using carbon dioxide. Finally, the samples were coated with 3 nm gold/palladium using a sputter coater (E5100 Polaron/Quorum Technologies, UK) and examined using a field emission SEM (Leo 1530 Gemini, Carl Zeiss Microscopy, Germany) with a 5 kV acceleration voltage and a working distance of 4.2 mm. Signals from an in-lens-SE and an Everhart-Thornley secondary electron detector were merged (50:50) for all samples.
Adherence to HEp-2 cells
HEp-2 epithelial cells were grown to 70–90% optical confluence in 24-well plates on glass coverslips (8 mm, Thomas Scientific, Germany) in DMEM FCS (with 0.45% glucose and 10% FCS; Lonza, Germany). Before adding bacteria, HEp-2 cells were washed three times with DMEM. Bacteria were grown overnight in LB broth, and 10 µl (~MOI of 100) were added to the HEp-2 cells with 1 ml DMEM containing 1% mannose and incubated for 3 h statically at 37 °C with 5% CO2. HEp-2 cells were washed five times with PBS, followed by fixation with 3% PFA for 20 min at room temperature. Samples were subsequently air dried and mounted for phase contrast microscopy at 1000-fold magnification on a Nikon Eclipse inverted microscope. For the quantitative HEp-2 adherence assay, 1 ml of PBS and 10 µl of 10% saponin was added after washing with PBS, followed by pipetting up and down approximately ten times, stepwise dilution in PBS and plating on LB agar to determine colony forming units (CFU). As reference for the amount of bacteria multiplied within the 3 h, the washing step with PBS was omitted and the counted CFUs was set to 100%.
Cytotoxicity assay
HEp-2 epithelial cells were grown to 70–90% optical confluence in 96-well plates in DMEM FCS (with 0.45% glucose and 10% FCS; Lonza, Germany). HEp-2 cells were washed three times with DMEM and then inoculated with bacteria cultured overnight (1:100) in DMEM containing 1% mannose (6 wells per strain). After 24 h, 50 µl of the supernatant was used with the CytoTox 96® Non-Radioactive Cytotoxicity Assay (Promega, Germany) to measure the relative cytotoxicity due to the release of lactate dehydrogenase (LDH) by leaky HEp-2 cells. Lysed HEp-2 cells without the addition of bacteria represented the maximum LDH release (100% cytotoxicity) according to manufacturer’s instructions.
Figure_S1
Download PDF (277.4 KB)Figure_S2
Download PDF (275.9 KB)Table S1
Download PDF (93.3 KB)Table S2
Download PDF (97.9 KB)Table S3
Download PDF (142.2 KB)Table S4
Download PDF (88.6 KB)Table S5
Download PDF (113.2 KB)Table S6
Download PDF (254.4 KB)Table S7
Download PDF (131.4 KB)Acknowledgements
We thank Rita Prager and Erhard Tietze of the Division of Enteropathogenic Bacteria and Legionella (Robert Koch-Institut, Wernigerode, Germany) for important suggestions and helpful discussions regarding the project, as well as Sangeeta Banerji for critical reading of the manuscript. We further acknowledge Ute Siewert, Ute Strutz, Thomas Garn and Karsten Großhenning for excellent technical assistance. We thank Jennifer Bender of the Division of Nosocomial Infectious Pathogens and Antibiotic Resistance (Robert Koch-Institut, Wernigerode, Germany) for assistance with Illumina MiSeq sequencing. The project was financially supported by the Intensified Molecular Surveillance Initiative of the Robert Koch Institute.
Author contributions
C.L.: conceived and designed the experiments, acquired data and conducted the experiments, analyzed the data, interpreted the results, drafted the manuscript, revised the manuscript, and approved the final version A.Fruth.: acquired data, interpreted the results, revised the manuscript, and approved the final version G.H.: conducted experiments, interpreted the results, revised the manuscript, and approved the final version M.L.: interpreted the results, revised the manuscript, and approved the final version S.M.: conducted experiments, interpreted the results, revised the manuscript, and approved the final version P.D.: interpreted the results, revised the manuscript, and approved the final version. A.Flieger: conceived and designed the experiments, interpreted the results, drafted the manuscript, revised the manuscript, and approved the final version
Conflict of interest
The authors declare that they have no conflicts of interest.
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41426-018-0209-8).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Robert-Koch-Institute. Final Presentation and Evaluation of the Epidemiological Findings in the EHEC O104:H4 Outbreak, Germany2011 (Robert-Koch-Institut, Berlin, Germany, 2011).
- FrankCet al.Epidemic profile of Shiga-toxin-producing Escherichia coli O104:H4 outbreak in GermanyNew Engl. J. Med.2011365 1771 178010.1056/NEJMoa1106483
- BielaszewskaMet al.Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological studyLancet Infect. Dis.20111167167610.1016/S1473-3099(11)70165-7
- MellmannAet al.Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technologyPLoS ONE20116e2275110.1371/journal.pone.00227513140518
- Navarro-Garcia, F. Escherichia coli O104:H4 pathogenesis: an enteroaggregative E. coli/Shiga toxin-producing E. coli explosive cocktail of high virulence. Microbiol. Spectr. 2, 10.1128/microbiolspec.EHEC-0008-2013 (2014).
- BoisenNMelton-CelsaARScheutzFO’BrienADNataroJPShiga toxin 2a and Enteroaggregative Escherichia coli--a deadly combinationGut Microbes2015627227810.1080/19490976.2015.10545914615819
- JohnsonTJNolanLKPathogenomics of the virulence plasmids of Escherichia coliMicrobiol Mol. Biol. Rev.20097375077410.1128/MMBR.00015-092786578
- MorabitoSet al.Enteroaggregative, Shiga toxin-producing Escherichia coli O111:H2 associated with an outbreak of hemolytic-uremic syndromeJ. Clin. Microbiol199836840842104641
- IyodaSet al.Inducible stx2 phages are lysogenized in the enteroaggregative and other phenotypic Escherichia coli O86:HNM isolated from patientsFEMS Microbiol Lett.200019171010.1111/j.1574-6968.2000.tb09311.x
- DallmanTet al.Characterization of a verocytotoxin-producing enteroaggregative Escherichia coli serogroup O111:H21 strain associated with a household outbreak in Northern IrelandJ. Clin. Microbiol2012504116411910.1128/JCM.02047-123502983
- MellmannAet al.Analysis of collection of hemolytic uremic syndrome-associated enterohemorrhagic Escherichia coliEmerg. Infect. Dis.2008141287129010.3201/eid1408.0710822600372
- TietzeEet al.Comparative genomic analysis of two novel sporadic Shiga toxin-producing Escherichia coli O104:H4 strains isolated 2011 in GermanyPLoS ONE201510e012207410.1371/journal.pone.01220744383531
- GradYHet al.Genomic epidemiology of the Escherichia coli O104:H4 outbreaks in Europe, 2011Proc. Natl Acad. Sci. USA20121093065307010.1073/pnas.1121491109
- Jourdan-da Silva, N. et al. Outbreak of haemolytic uraemic syndrome due to Shiga toxin-producing Escherichia coli O104:H4 among French tourists returning from Turkey, September 2011. Euro Surveill.17, 20065 (2012).
- MoneckeSet al.Presence of enterohemorrhagic Escherichia coli ST678/O104:H4 in France prior to 2011Appl. Environ. Microbiol.2011778784878610.1128/AEM.06524-113233108
- AhmedSAet al.Genomic comparison of Escherichia coli O104:H4 isolates from 2009 and 2011 reveals plasmid, and prophage heterogeneity, including shiga toxin encoding phage stx2PLoS ONE20127e4822810.1371/journal.pone.00482283486847
- GuyLet al.Genomic diversity of the 2011 European outbreaks of Escherichia coli O104:H4Proc. Natl Acad. Sci. USA2012109E3627E362810.1073/pnas.1206246110
- SchmidtHet al.Development of PCR for screening of enteroaggregative Escherichia coliJ. Clin. Microbiol199533701705228017
- PragerRet al.Two novel EHEC/EAEC hybrid strains isolated from human infectionsPLoS ONE20149e9537910.1371/journal.pone.00953793994036
- OkekeINNataroJPEnteroaggregative Escherichia coliLancet Infect. Dis.2001130431310.1016/S1473-3099(01)00144-X
- KaperJBNataroJPMobleyHLPathogenic Escherichia coliNat. Rev. Microbiol20042123110.1038/nrmicro818
- TokudaKet al.Characterization of typical and atypical enteroaggregative escherichia coli in Kagoshima, Japan: biofilm formation and acid resistanceMicrobiol Immunol.20105432032910.1111/j.1348-0421.2010.00210.x
- JonssonRet al.Novel aggregative adherence fimbria variant of enteroaggregative Escherichia coliInfect. Immun.2015831396140510.1128/IAI.02820-144363450
- MonteroDAet al.Locus of Adhesion and Autoaggregation (LAA), a pathogenicity island present in emerging Shiga Toxin-producing Escherichia coli strainsSci. Rep.2017710.1038/s41598-017-06999-y5539235
- Centers for Disease Control and Prevention (CDC). National STEC Surveillance Annual report, 2014. Atlanta, Georgia: US Department of Health and Human Services, CDC, 2017.
- AndradeFBGomesTATEliasWPA sensitive and specific molecular tool for detection of both typical and atypical enteroaggregative Escherichia coliJ. Microbiol Methods2014106161810.1016/j.mimet.2014.07.030
- LimaIFet al.Prevalence of enteroaggregative Escherichia coli and its virulence-related genes in a case-control study among children from north-eastern BrazilJ. Med. Microbiol.20136268369310.1099/jmm.0.054262-03709657
- SrimanotePPatonAWPatonJCCharacterization of a novel type IV pilus locus encoded on the large plasmid of locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humansInfect. Immun.2002703094310010.1128/IAI.70.6.3094-3100.2002128018
- ClarkeSCHaighRDFreestonePPWilliamsPHVirulence of enteropathogenic Escherichia coli, a global pathogenClin. Microbiol Rev.20031636537810.1128/CMR.16.3.365-378.2003164217
- NataroJPMaherKOMackiePKaperJBCharacterization of plasmids encoding the adherence factor of enteropathogenic Escherichia coliInfect. Immun.19875523702377260715
- TobeTet al.Complete DNA sequence and structural analysis of the enteropathogenic Escherichia coli adherence factor plasmidInfect. Immun.1999675455546296904
- GunzburgSTTornieporthNGRileyLWIdentification of enteropathogenic Escherichia coli by PCR-based detection of the bundle-forming pilus geneJ. Clin. Microbiol19953313751377228170
- Hebbelstrup JensenBOlsenKEStruveCKrogfeltKAPetersenAMEpidemiology and clinical manifestations of enteroaggregative Escherichia coliClin. Microbiol Rev.20142761463010.1128/CMR.00112-134135892
- HarringtonSMStraumanMCAbeCMNataroJPAggregative adherence fimbriae contribute to the inflammatory response of epithelial cells infected with enteroaggregative Escherichia coliCell Microbiol200571565157810.1111/j.1462-5822.2005.00588.x
- MeloARet al.Expression of the virulence factor, BfpA, by enteropathogenic Escherichia coli is essential for apoptosis signalling but not for NF-kappaB activation in host cellsScand. J. Immunol.20056151151910.1111/j.1365-3083.2005.01626.x
- DallmanTJet al.An investigation of the diversity of strains of enteroaggregative Escherichia coli isolated from cases associated with a large multi-pathogen foodborne outbreak in the UKPLoS ONE20149e9810310.1371/journal.pone.00981034028294
- Basso, P. et al. Pseudomonas aeruginosa pore-forming exolysin and type iv pili cooperate to induce host cell lysis. mBio8, 10.1128/mBio.02250-16 (2017).
- FullerCAPellinoCAFlaglerMJStrasserJEWeissAAShiga toxin subtypes display dramatic differences in potencyInfect. Immun.2011791329133710.1128/IAI.01182-103067513
- KarveSSWeissAAGlycolipid binding preferences of Shiga toxin variantsPLoS ONE20149e10117310.1371/journal.pone.01011734077739
- FurukawaTet al.Fatal hemorrhage induced by subtilase cytotoxin from Shiga-toxigenic Escherichia coliMicrob. Pathog.20115015916710.1016/j.micpath.2011.01.0023385872
- YahiroKMorinagaNMossJNodaMSubtilase cytotoxin induces apoptosis in HeLa cells by mitochondrial permeabilization via activation of Bax/Bak, independent of C/EBF-homologue protein (CHOP), Ire1alpha or JNK signalingMicrob. Pathog.20104915316310.1016/j.micpath.2010.05.0073417112
- SavarinoSJet al.Enteroaggregative Escherichia coli heat-stable enterotoxin 1 represents another subfamily of E. coli heat-stable toxinProc. Natl Acad. Sci. USA1993903093309710.1073/pnas.90.7.3093
- TanAet al.Evolutionary adaptation of an AraC-like regulatory protein in Citrobacter rodentium and Escherichia speciesInfect. Immun.2015831384139510.1128/IAI.02697-144363417
- MorinNSantiagoAEErnstRKGuillotSJNataroJPCharacterization of the AggR regulon in enteroaggregative Escherichia coliInfect. Immun.20138112213210.1128/IAI.00676-123536136
- Martinez-LagunaYCalvaEPuenteJLAutoactivation and environmental regulation of bfpT expression, the gene coding for the transcriptional activator of bfpA in enteropathogenic Escherichia coliMol. Microbiol19993315316610.1046/j.1365-2958.1999.01460.x
- DudleyEGThomsonNRParkhillJMorinNPNataroJPProteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coliMol. Microbiol2006611267128210.1111/j.1365-2958.2006.05281.x
- NataroJPYikangDYingkangDWalkerKAggR, a transcriptional activator of aggregative adherence fimbria I expression in enteroaggregative Escherichia coliJ. Bacteriol.19941764691469910.1128/jb.176.15.4691-4699.1994196291
- IguchiAet al.Complete genome sequence and comparative genome analysis of enteropathogenic Escherichia coli O127:H6 strain E2348/69J. Bacteriol.200919134735410.1128/JB.01238-08
- ChaudhuriRRet al.Complete genome sequence and comparative metabolic profiling of the prototypical enteroaggregative Escherichia coli strain 042PLoS ONE20105e880110.1371/journal.pone.00088012808357
- ScheutzFet al.Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclatureJ. Clin. Microbiol2012502951296310.1128/JCM.00860-123421821
- FruthAPragerRTietzeERabschWFliegerAMolecular epidemiological view on Shiga toxin-producing Escherichia coli causing human disease in Germany: Diversity, prevalence, and outbreaksInt J. Med Microbiol201530569770410.1016/j.ijmm.2015.08.020
- Halbedel, S. et al. Whole genome sequencing of recent Listeria monocytogenes isolates from Germany reveals population structure and disease clusters. J. Clin. Microbiol, 10.1128/JCM.00119-18 (2018).
- NascimentoMet al.PHYLOViZ 2.0: providing scalable data integration and visualization for multiple phylogenetic inference methodsBioinformatics20173312812910.1093/bioinformatics/btw582
- FramptonMHoulstonRGeneration of artificial FASTQ files to evaluate the performance of next-generation sequencing pipelinesPLoS ONE20127e4911010.1371/journal.pone.00491103495771
- DatsenkoKAWannerBLOne-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR productsProc. Natl Acad. Sci. USA2000976640664510.1073/pnas.120163297
- BanerjiSBewersdorffMHermesBCianciottoNPFliegerACharacterization of the major secreted zinc metalloprotease- dependent glycerophospholipid:cholesterol acyltransferase, PlaC, of Legionella pneumophilaInfect. Immun.2005732899290910.1128/IAI.73.5.2899-2909.20051087370
- GibsonDGet al.Enzymatic assembly of DNA molecules up to several hundred kilobasesNat. Methods2009634334510.1038/nmeth.1318
- LaemmliUKCleavage of structural proteins during the assembly of the head of bacteriophage T4Nature197022768068510.1038/227680a05432063
- LevineMMet al.Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasiveLancet197811119112210.1016/S0140-6736(78)90299-4
- AppleyardRKSegregation of new lysogenic types during growth of a doubly lysogenic strain derived from Escherichia Coli K12Genetics1954394404521209664
