 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
The funnel plot is a graphical visualization of summary data estimates from a meta-analysis, and is a useful tool for detecting departures from the standard modeling assumptions. Although perhaps not widely appreciated, a simple extension of the funnel plot can help to facilitate an intuitive interpretation of the mathematics underlying a meta-analysis at a more fundamental level, by equating it to determining the center of mass of a physical system. We used this analogy to explain the concepts of weighing evidence and of biased evidence to a young audience at the Cambridge Science Festival, without recourse to precise definitions or statistical formulas and with a little help from Sherlock Holmes! Following on from the science fair, we have developed an interactive web-application (named the Meta-Analyser) to bring these ideas to a wider audience. We envisage that our application will be a useful tool for researchers when interpreting their data. First, to facilitate a simple understanding of fixed and random effects modeling approaches; second, to assess the importance of outliers; and third, to show the impact of adjusting for small study bias. This final aim is realized by introducing a novel graphical interpretation of the well-known method of Egger regression.
1. Introduction
Meta-analysis is the science of combining the evidence available from different sources to arrive at the most sensible and informed conclusion to a given question. The technique has a long history in medical research, where it is routinely used to aggregate results from independent clinical trials testing competing therapies (e.g., which is better: treatment A or B? And by how much?). While each study may be too small or imprecise to definitively answer the clinical question on its own, the overall estimate delivered by a meta-analysis can often do so, thus negating the need to perform further costly research. Its application is not limited to medicine, and spans the entire scientific spectrum. For example, it has been used to synthesize studies measuring: aspects of human psychology such as emotion or motivation (Elfenbein and Ambady Citation2002); the performance of standard equipment across laboratories (Paule and Mandel Citation1982); and the rest mass of elementary particles in physics (Baker and Jackson Citation2013).
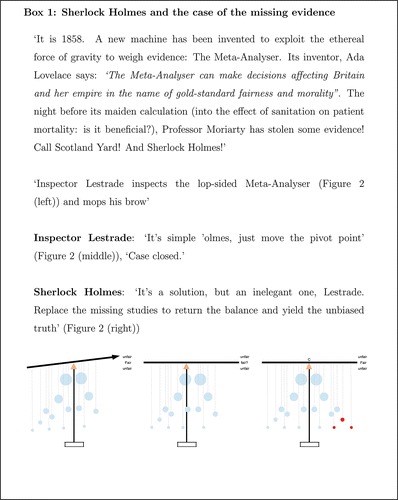
Scientists tasked with performing a meta-analysis are often hindered by the suspicion that the set of studies they have collected form an incomplete and biased selection of the total evidence base. One reason why this may occur is that a study's outcome (or result) can affect its chances of being published and put into the public domain (Rosenthal Citation1979). This can, in turn, cause authors to selectively report study results to increase their chances of being published (Ioannidis Citation2005; Bowden, Jackson, and Thompson Citation2010). Therefore, bias can be due as much to studies that are missing as to those that are present. In 2014, we attempted to explain the notion of meta-analysis and the concept of biased evidence to a young audience at the Cambridge Science Festival (http://www.cam.ac.uk/sciencefestival/), an event for Cambridge academics working across the scientific spectrum to promote and explain their current research to the general public. To achieve our specific aim without recourse to mathematical formulas, or any precise definitions of bias, we came up with a mechanical device, named the “Meta-Analyser.” We introduced our invention within the context of a Sherlock Holmes inspired mystery—see Box 1 for an explanation and for pictures of the Meta-Analyser in action at the festival.
The Meta-Analyser shown in and is a machine that can be used to find the balance or pivot point of a set of weights by hanging the weights from a pole in such a way that the pole is parallel to the ground. It was explained to the children that the weights reflected the importance of each source of information in the calculation.
Figure 2. Left: Unbalanced Meta-Analyser. Middle: Rebalanced Meta-Analyser (Inspector Lestrade approach). Right: Rebalanced Meta-Analyser (Sherlock Holmes approach).
Although it does not materially affect the balance point, the length of cord connecting each weight to the pole was chosen to be proportional to its weight. With this convention in place, the Meta-Analyser can be interpreted as a glorified Funnel plot (Sterne and Egger Citation2001). Funnel plots are used in meta-analysis to visually assess whether the data are potentially affected by bias. If the study results are not symmetrical about the pivot point (or funnel shaped) then missing studies could be responsible.
By augmenting the Funnel plot with an additional pole, cord, and pivot, the Meta-Analyser gives this abstract object, and the overall estimate that it implies, a clear physical interpretation. It also facilitates a simple, graphical explanation of the general notion of biased evidence. Box 1 shows the story as explained to the (mainly young) science fair attendees: A small number of study results were removed, leading to an imbalance in the Meta-Analyser. Inspector Lestrade's typically lacklustre approach is to ignore the missing information altogether, and recalculate the pivot point. Sherlock Holmes’ solution is to add in “missing” studies to rebalance and restore symmetry to the Meta-Analyser. Aficionados will notice this as a simplistic representation of the “Trim and Fill” method Duval and Tweedie (Citation2000).
The Meta-Analyser was intended to be a simple educational tool to demonstrate the basics of evidence synthesis to a lay audience, capitalizing on the growing recognition of Ada Lovelace in the birth of computation through initiatives such as Ada Lovelace Day (http://findingada.com/) and jumping on the bandwagon of “steampunk” fiction, which melds Victorian fantasy and futurism (Onion Citation2008; Tanenbaum, Tanenbaum, and Wakkary Citation2012). However, we continued to find the physical system it describes useful, more broadly, to explain some common secondary issues in meta-analysis. We have therefore developed an online web application (the Meta-Analyser) to implement the approach in practice and to bring this idea to a wider audience of students and researchers. Moreover, our work revealed that the physical analogy the Meta-Analyser promotes is perhaps a deeper and more accurate description of the mathematics underlying a meta-analysis than many statisticians realize.
In Section 2, we give a more formal introduction to meta-analysis and introduce the Meta-Analyser web app, showing that it helps to transparently demonstrate the implications of moving from a fixed to a random effects model and to assess the influence of outlying studies. In Section 3, we discuss the issue of biased evidence and how small study bias is commonly addressed by medical statisticians (the authors’ field) using Egger regression (Egger et al. Citation1997). We then show that by viewing Egger regression from an estimating equation perspective, a novel causal interpretation of this method is found that can be intuitively visualized via the Meta-Analyser. We conclude with a discussion of the issues raised in Section 4 and point to future research. Sections 3.1 and 4.3 formulate previously explained ideas using the estimating equation framework. They may be of most interest to the statistically inclined reader, but can be safely ignored by those wishing to focus on the graphics content of this article alone.
2. The Meta-Analyser As a Research Tool
2.1. A First Statistical Model for Meta-Analysis
Let yi, i = 1, …, k, represent summary estimates of the same apparent quantity from k independent information sources in a meta-analysis. The ith estimate is associated with a variance, s2i that is assumed to be known. The standard fixed effect model for combining the yis assumes
(1)
(1) and the focus for inference is the population mean effect parameter μ. If si is uncorrelated with εi in Equation (Equation1
(1)
(1) ), so that E[siεi] = 0, then each yi is an unbiased estimate for μ. The fixed effect estimate
and its variance are given by the well-known formulas
(2)
(2) where the weight given to study i is inversely proportional to its variance. This weighting is attractive, since it minimizes the variance of the overall estimate,
. The Meta-Analyser (https://chjackson.shinyapps.io/MetaAnalyser/) is an online free-to-access tool to visualize the results from a meta-analysis using the physical analogy previously described. shows the Meta-Analyser populated with a fictional collection of k = 13 studies. This is the “Symmetric” example dataset included in our tool. It shows a standard funnel plot (Sterne and Egger Citation2001) that has been augmented so that the area representing point i is proportional to wi = 1/s2i, to promote its interpretation as a physical mass. Points are joined by vertical cord to a pole that is joined itself to a vertical stand at a pivot point, p. Study weights are imagined to exert a downward force due to gravity. Since the pole is perfectly horizontal, it is intuitively understood that p satisfies the physical law:
(3)
(3) and is therefore equal to the center of mass (Beatty Citation2005). Formula (Equation3
(3)
(3) ) can be viewed as a rudimentary estimating equation, a construction we continue to use throughout this article. It is simple to verify that solving for p yields the fixed effect estimate
in (Equation2
(2)
(2) ). The length of the stand base along the x-axis shows the 95% confidence interval for the overall effect μ, which is approximately (−0.75,0.75).
Figure 3. The Meta-Analyser supporting the symmetric data, a fictional body of 13 study results. The center of mass (overall estimate) is located at zero.
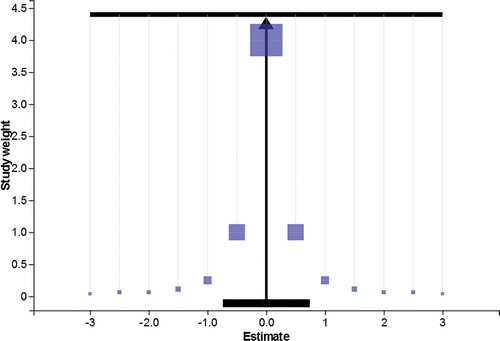
If the modeling assumption E[siεi] = 0 is true, this would imply that study effect sizes and precisions are also uncorrelated. The Meta-Analyser should then appear symmetrical, in that less precise estimates should funnel in from either side toward the most precise estimates. The Meta-Analyser therefore provides a simple means to visually assess the reliability of the overall estimate . Note that this is exactly satisfied for the toy example in . When populated with such idealized data, the Meta-Analyser may remind some of Galton's bean board or quincunx, a tool that also illustrates how statistical laws can emerge from a physical process in the presence of gravity.
Humans tend to be good at perceiving lengths in graphs, but usually under-estimate areas, and do even worse with volumes (Cleveland and McGill Citation1984). However, as a reviewer commented, the Meta-Analyser is not relying solely on areas, but rather the entire system in balance and the ability to interact. The Meta-Analyser gives users the flexibility to show the weights in absolute (wi = 1/s2i) or percentage (wi/∑wi) terms and to change the scale of the y-axis to improve its aesthetic quality. This is particularly useful for those wishing to include an image of the Meta-Analyser graphical output in a separate document, as done here.
3. Random Effects Models
In a fixed effect meta-analysis, all studies are assumed to provide an estimate of the same quantity, and therefore any differences between study estimates should be due to chance variation alone. If the fixed effect model is true, then we would expect Cochran's Q-statistic, Q = to be approximately equal to k − 1. If, as is often the case, Q is significantly larger than k − 1, then the fixed effect model is thought implausible. A common secondary issue in meta-analysis, therefore, is accounting for between study heterogeneity via extended modeling approaches.
Two distinct methods for incorporating heterogeneity have emerged. The first is via an additional additive random effect, as in model (Equation4(4)
(4) ) (e.g., DerSimonian and Laird Citation1986; Higgins and Thompson Citation2002). The second is via the addition of a multiplicative scale factor, as in model (Equation5
(5)
(5) ) (e.g., Thompson and Sharp Citation1999; Baker and Jackson Citation2013):
(4)
(4)
(5)
(5) The point estimate for μ obtained by fitting model (Equation5
(5)
(5) ) is always identical to that obtained from fitting model (Equation1
(1)
(1) ), since the common term φ simply cancels from the numerator and denominator in (Equation2
(2)
(2) ). Only the variance of the estimate is altered, due to the weight given to each study being rescaled by a factor of φ (when φ is not equal to one). Consequently, when the study weights are presented in percentage terms, moving from a fixed effect model to a multiplicative random effects model does not induce any physical change in the Meta-Analyser at all. In contrast, both the point estimate and variance for
change under the additive random effect model (Equation4
(4)
(4) ) when τ2 is estimated to be greater than zero. Random effects model (Equation4
(4)
(4) ) is popular in medical research; it is promoted by the Cochrane collaboration (Higgins and Green Citation2011) who are responsible for a large proportion of all meta-analyses in bio-medicine. Moreover, since its implementation via the Meta-Analyser demands a significant graphical extension (as discussed in Section 3.2), we focus on model (Equation4
(4)
(4) ) for the remainder of this section. The assumption E[siδiεi] = 0 implicit in model (Equation4
(4)
(4) ) is required to ensure that each study still provides an unbiased estimate for μ.
It is common practice to estimate τ2 under model (Equation4(4)
(4) ) using the simple formula defined by DerSimonian and Laird (Citation1986), to give
. This estimate conveniently provides a link between Q and a popular measure of heterogeneity, I2 (Higgins and Thompson Citation2002) as follows:
where s2 approximates the typical within study variance. When Q is less than k − 1,
is simply set to zero. Estimation of μ (and its variance) then follows by simple application of formula (Equation2
(2)
(2) ) except now the weight given to study i, wi, changes to
.
3.1. An Estimating Equation Formulation of Model (Equation4 (4)
(4)  (4) )
(4) )
To continue to provide a physical interpretation of the calculations underpinning a meta-analysis, and as a natural progression of the single estimating equation for fixed effect model (Equation1(1)
(1) ), we can formulate random effects model (Equation4
(4)
(4) ) via a system of estimating equations as follows:
(6)
(6)
(7)
(7)
(8)
(8)
Formula (Equation8(8)
(8) ) is referred to as the generalized Q statistic (Bowden et al. Citation2011) and, when solved in conjunction with (Equation6
(6)
(6) ) and (Equation7
(7)
(7) ), it returns the Paule–Mandel (PM) estimate for τ2 (Paule and Mandel Citation1982), which we denote by
. As with
the PM estimate is constrained to be positive, and it is known to provide a more reliable estimate for the between study heterogeneity than
(Veroniki et al. Citation2016).
3.2. Random Effects Models via the Meta-Analyser
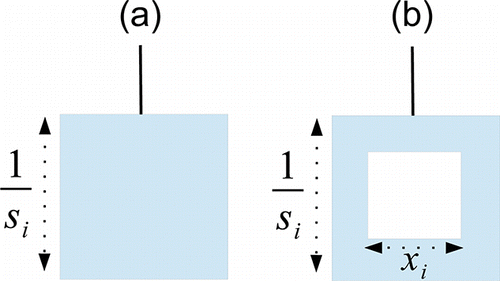
Application of the random effects model, with additional variance component τ2, leads to study results being both down-weighted and more similarly weighted. Furthermore, the original weight given to large studies is reduced to a greater extent than those of smaller studies. This issue is fairly subtle and hard to comprehend, but the Meta-Analyser provides a very simple visualization, by directly adjusting the mass of each weight. Specifically, we represent the weight “loss” induced by moving from a fixed to an additive random effects model by “drilling out” a square of length xi to satisfy the Pythagorean identity =
, as illustrated in .
Figure 4. (a) Weight given to study i in a fixed effect meta-analysis. (b) Weight given to study i in an additive random effects meta-analysis. A square of length xi is removed from weight i to satisfy the Pythagorean formula.
(left) shows the Meta-Analyser populated with a dataset of eight randomized trial results, each assessing the use of magnesium to treat myocardial infarction. The effect measure, yi, is the log-odds ratio of death between treatment and control groups for study i. These data were previously analyzed by Higgins and Spiegelhalter (Citation2002). Between trial heterogeneity is present for these data ( = 0.095, I2 = 27.6%), so our analysis is under random effects model (Equation4
(4)
(4) ) using
to estimate τ2. Full results obtained via the estimating equation approach (and so using the PM estimate for τ2) are shown in . Under the random effects model, wi is reduced from
to 1/(s2i +
). The center of mass or balance point defined by the holed-out weights in (left) is automatically consistent with the random effects estimate for μ. It is immediately apparent that large studies lose a higher proportion of their fixed effect weight than small studies under this model.
Figure 5. The Meta-Analyser supporting the magnesium data under a random effects model for all trials (left); and with the Shechter trial removed (right).
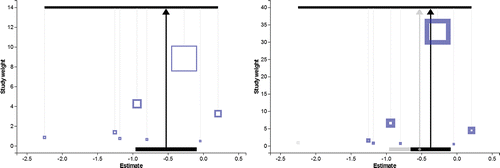
Table 1. Meta-analysis of the magnesium data under random effects model (Equation3(3)
(3) ), with and without the Shechter trial. S.E. = standard error.
3.3. Outliers and Sensitivity Analysis
The amount of heterogeneity estimated in a meta-analysis can depend heavily on extreme, and often small, study results (Bowden et al. Citation2011). It is therefore useful in some circumstances to perform a sensitivity analysis, in which an outlying study result is excluded. (right) shows the Meta-Analyser supporting the magnesium data with the Shechter study (shown in gray) excluded. This is achieved by simply clicking on its weight. The solid black support stand shows the overall estimate and corresponding 95% confidence interval in this case. For comparison, the original point estimate and confidence interval using all the data is kept but shown in gray. In this example, exclusion of the outlying study removes a large proportion of the between trial heterogeneity (updated = 0.012, I2 = 5.2%). From looking at the scale of the y-axis, one can see that the weight given to each study has increased, with the largest study benefitting the most. The Meta-Analyser facilitates easy transitions between various models like this as part of a sensitivity analysis. Users also see the Meta-Analyser dynamically tip and rebalance in response to the latest analysis choice.
4. Small Study Bias
4.1. The Aspirin Data
(left) shows the Meta-Analyser enacted on 63 randomized controlled trials reported by Edwards et al. (Citation2000) that each investigated the benefit of oral Aspirin for pain relief. Study estimates yi represent the log-odds ratios for the proportion of patients in each arm who had at least a 50% reduction in pain. Between trial heterogeneity was present for these data ( = 0.04, I2 = 10%) and (left) reflects the weight given toeach study by the Meta-Analyser under the random effects model (Equation4
(4)
(4) ) using
as the heterogeneity parameter estimate. Despite the apparent between trial heterogeneity, the conclusion of the random effects meta-analysis is that oral Aspirin is an effective treatment, the combined log-odds ratio estimate is 1.26 in favor of Aspirin with a 95% confidence interval (1.1,1.41). Full results are shown in .
Figure 6. Left: The Meta-Analyser supporting the Aspirin data under a random effects model. Right: The Meta-Analyser incorporating Egger regression enacted on the Aspirin data and shown under the potential outcome transform.
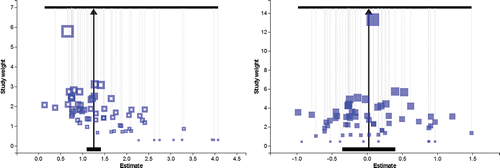
Table 2. Results for Meta-analyses of the Aspirin data. S.E. = standard error.
The hypothetical data shown in is perfectly symmetrical about its center of mass, indicating that there is no correlation between effect size and precision across studies. However, there is a clear asymmetry present in the Aspirin data, smaller studies tend to show larger effect size estimates, whereas larger studies tend to report more modest results. For these data, Cor(yi, 1/si) = −0.7, which suggests that E[siδiεi] = 0 does not hold for model (Equation4(4)
(4) ). The phenomenon of observing a negative correlation between study precision and effect size is often given the umbrella term “small study bias” (Egger et al. Citation1997; Rücker et al. Citation2011; Sterne et al. Citation2011).
Small study bias could actually be caused by real differences between small and large studies. Small trials may employ a more intensive intervention and therefore generate a greater effect on disease outcomes than larger trials (Egger et al. Citation1997; Bowater and Escarela Citation2013). Asymmetry could also be a simple artefact of the data. For example, point estimates are not strictly independent of their estimated variances when calculated from binary or count outcomes, (Harbord, Egger, and Sterne Citation2006, Peters et al. Citation2010). However, its cause could also be more sinister. Publication bias, or the file-drawer problem (Rosenthal Citation1979) occurs when journals selectively publish study results that achieve a high level of statistical significance, and also induces asymmetry.
4.2. Egger Regression
When attempting to explain the concept of biased evidence and of bias adjustment to the science festival audience, we opted for a simplified version of Trim and Fill (Duval and Tweedie Citation2000). Trim and Fill can be essentially understood as follows. First, smaller studies causing the asymmetry are removed (or “trimmed”) from the data, and the remaining studies are used to estimate the overall effect. Then the trimmed studies are replaced (or “filled”), along with their mirror images, by reflecting them about vertical line defined by the overall effect estimate. Trim and Fill therefore provides an estimate for the number of missing studies and a bias-adjusted estimate for the overall effect. Because of its intuitive nature and visual appeal, it was natural to use this idea within our Sherlock Holmes mystery (Box 1). However, the mathematics behind Trim and Fill are quite complicated and some dislike the way its results depend on imputed data. Perhaps due to its relative simplicity, the most popular approach to testing and adjusting for small study bias in medical research is Egger regression (Egger et al. Citation1997). This assumes the following linear fixed effect model to explain the correlation between yi and 1/si:
(9)
(9) The addition of an intercept parameter β0 in model (Equation9
(9)
(9) ) allows small study bias to be accounted for. Testing for small study bias is then equivalent to testing H0: β0 = 0. If E[siεi] = 0 for model (Equation9
(9)
(9) ), then the overall effect estimate,
, adjusted for possible small study bias (via
) is a consistent estimate for μ. Several authors have considered the addition of random effects into model (Equation9
(9)
(9) ), to account for possible residual heterogeneity after adjustment for small study bias, see, for example, Moreno et al. (Citation2009), Peters et al. (Citation2010), and Rücker et al. (Citation2011). Their approaches have been straightforward generalizations of the additive and multiplicative random effects models (Equation4
(4)
(4) ) and (Equation5
(5)
(5) ), respectively, as below:
(10)
(10)
(11)
(11) At first sight, model (Equation10
(10)
(10) ) seems the most natural extension to model (Equation9
(9)
(9) ). However, when a nonzero value for τ2 is estimated under model (Equation10
(10)
(10) ), the resulting overall estimate for μ differs from, and can often exhibit more substantial bias than the fixed effect estimate (Rücker et al. Citation2011). By contrast, multiplicative model (Equation11
(11)
(11) ) is far more well behaved in this respect since its point estimate is identical to that obtained from fitting model (Equation9
(9)
(9) ). Nullifying the influence of variance components on the overall mean, a property enjoyed by model (Equation11
(11)
(11) ) is so attractive in the presence of small study bias, that approaches have been developed to artificially incorporate this feature into additive random effects models as well (Henmi and Copas Citation2010).
For these reasons, we analyze the Aspirin data using the multiplicative Egger regression model only. This is straightforward, because model (Equation11(11)
(11) ) is automatically fitted by the process of standard linear regression, in which the variance of the residual error is estimated, rather than assumed to be 1. For these data,
= 2.11, with a p-value of approximately 1 × 10− 8,
is equal to 0.025, with a p-value of 0.89, and
= 0.64. In summary, Egger regression detects a highly significant presence of small study bias and, after this has been removed, no evidence of a treatment effect whatsoever.
4.3. An Estimating Equation Formulation of Model (Equation11 (11)
(11)  (11) )
(11) )
We now show that, like Trim and Fill, Egger regression can be understood as a means to de-bias a meta-analysis by restoring symmetry to the funnel plot, in a way that complements its application via the Meta-Analyser and makes connections with the world of causal inference. We start by assuming model (Equation11(11)
(11) ). Multiplying each side by si and subtracting β0si yields
The term yi(β0) is a transformed version of the effect size estimates, and when E[siεi] = 0, yi(β0) is mean-independent of si. The intercept estimate obtained from fitting model (Equation11(11)
(11) ),
, can be viewed as defining a particular transform of the data that forces
to be independent of si across all studies. We note the similarity between this formulation of Egger regression and the structural mean model framework in causal inference, in which observed outcomes are related to potential outcomes (Rubin Citation2005) using a parametric transform to satisfy statistical independence rules. Parameter estimation via this strategy is termed G-estimation, and has been used extensively to adjust for biases due to noncompliance or dropout in clinical trials and confounding bias in observational studies. For a general overview, see Vansteelandt and Joffe (Citation2014) or, in the context of epidemiology, Bowden and Vansteelandt (Citation2011). For this reason, we refer to yi(β0) as the potential outcome transform. Employing the estimating equation framework, model (Equation11
(11)
(11) ) is equivalent to solving the following system:
(12)
(12)
(13)
(13)
(14)
(14) where
is the arithmetic mean of the si terms. We now clarify the connection between the above system of estimating equations and estimation of β0, μ, and φ using standard linear regression theory. Fitting model (Equation9
(9)
(9) ) to obtain estimates for β0 and μ is equivalent to solving Equations (Equation12
(12)
(12) ) and (Equation13
(13)
(13) ) leaving φ unspecified (in the Appendix we provide some simple R code to verify this). We then formally define φ as a parameter and solve equation (Equation14
(14)
(14) ) to give
(15)
(15)
We note the equivalence of the numerator of Equation (Equation15(15)
(15) ) to the Q
statistic defined in Rücker et al. (Citation2011). The variances of
and
are given by
where
is the sample mean of the 1/s2i terms.
Concerns over the use of Egger regression have been raised when analyzing binary data because a study's outcome estimate will not truly be independent of its standard error, even when small study bias is not present. For this reason, Peters et al. (Citation2010) proposed to replace si in the Egger regression equation with a measure of precision based on study size, 1/ni say. This could easily be represented in our estimating equation with suitable modification. For example, G-estimation Equation (Equation13(13)
(13) ) above would then be used to find the β0 that forces independence between yi(β0) = yi − β0/ni and 1/ni.
4.4. Reanalysis of the Aspirin Data via the Meta-Analyser
Returning to the Aspirin data, (right) shows the final resting point of the Meta-Analyser upon enacting Egger regression using the estimating equation interpretation described above. Once the Egger regression option is ticked, users observe a dynamical change in the Meta-Analyser from its initial starting point of the standard random effects analysis in (left). The x-axis position is now the transformed or potential outcome scale = yi(2.11) for study i. The meta-analysis now exhibits a high amount of symmetry that can be immediately visualized by the user. This transformation is highlighted in , which plots potential outcomes
on the horizontal versus
on the vertical axis with the original data (yi vs. 1/si) also shown for comparison. When small study bias is present, estimates from small studies are shifted by a relatively large amount in the horizontal dimension (in this case to the left), whereas those from large studies are shifted horizontally by a relatively small amount.
Figure 7. Funnel plot of the original Aspirin data (yi vs. 1/si, hollow red dots) versus its transformed counterpart ( vs.
, solid black dots).
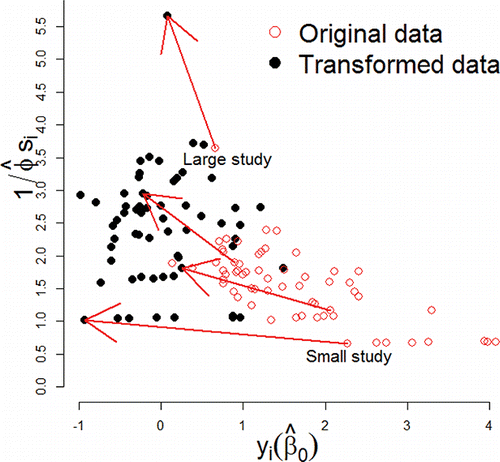
Because φ is estimated to be 0.64 in this instance, indicating under-dispersion after adjustment for small study bias, study precisions under the potential outcome transformation are increased in proportion to their size, so that the precisions of small studies are shifted vertically upward by a small amount and those of large studies are shifted by a large amount. If over-dispersion had been observed after adjustment, we would have seen a shift vertically downward instead.
5. Discussion
From its original conception and success as a science festival exhibit aimed at a lay audience, we believe that the Meta-Analyser will prove a useful online tool for explaining the rationale, and interpreting the effect of, extended modeling choices in meta-analysis to a more advanced audience. It has also proved a useful vehicle for forging new connections between methods of bias adjustment from the statistical literature.
To keep the Meta-Analyser as faithful as possible to the original funnel plot, the length of the cord connecting each weight to the horizontal pole was also set to be proportional to the weight. Clearly, only the weight matters for the physical interpretation. This redundancy could be exploited to come up with different versions of the Meta-Analyser. For example, the cord lengths could all be uniform, but this would lose the ability to detect asymmetry. One could also modify the weights. At present they are holed out when between study heterogeneity is present, to show how the weight of each study decreases. Other ways of achieving the same end are surely possible. The program code for the Meta-Analyser is available on the GitHub repository https://github.com/chjackson/MetaAnalyser. An R package containing the Meta-Analyser is also available, which allows users to run the application off-line. This can be installed from the repository as follows:
install.packages(''devtools'') # if necessary
devtools::install_github(''rstudio/DT'')
devtools::install_github(''chjackson/MetaAnalyser'')
We hope to continue to develop and improve the Meta-Analyser to incorporate new features and analysis choices. New ideas and offers of collaboration are very welcome.
Acknowledgments
The authors are grateful to the reviewers whose comments enabled them to greatly improve this manuscript. The authors thank the MRC visual aids department for their help in building the Meta-Analyser, the MRC Biostatistics Unit staff members who demonstrated its use at the Science fair (Simon White and Vikki O’neill in particular), and Anne Presanis for providing the photographs used in . The authors also thank Dan Jackson and George Davey Smith for their helpful comments on an early draft of this work.
Funding
Dr Jack Bowden is supported by an MRC methodology research fellowship (grant number MR/N501906/1).
References
- Baker, R., and Jackson, D. (2013), “Meta-Analysis Inside and Outside Particle Physics: Two Traditions That Should Converge?” Research Synthesis Methods, 4, 109–124.
- Beatty, M. (2005), Principles of Engineering Mechanics, Volume 2: Dynamics: The Analysis of Motion, New York: Springer Science & Business Media.
- Bowater, R., and Escarela, G. (2013), “Heterogeneity and Study Size in Random-Effects Meta-Analysis,” Journal of Applied Statistics, 40, 2–16.
- Bowden, J., Jackson, D., and Thompson, S. G. (2010), “Modelling Multiple Sources of Dissemination Bias in Meta-Analysis,” Statistics in Medicine, 29, 945–955.
- Bowden, J., Tierney, J., Copas, A., and Burdett, S. (2011), “Quantifying, Displaying and Accounting for Heterogeneity in the Meta-Analysis of RCTs Using Standard and Generalised Q Statistics,” Medical Research Methodology, 11, 41.
- Bowden, J., and Vansteelandt, S. (2011), “Mendelian Randomization Analysis of Case-Control Data Using Structural Mean Models,” Statistics in Medicine, 30, 678–694.
- Cleveland, W. S., and McGill, R. (1984), “Graphical Perception: Theory, Experimentation, and Application to the Development of Graphical Methods,” Journal of the American Statistical Association, 79, 531–554.
- DerSimonian, R., and Laird, N. (1986), “Meta-Analysis in Clinical Trials,” Controlled Clinical Trials, 7, 177–188.
- Duval, S., and Tweedie, R (2000), “Trim and Fill: A Simple Funnel-Plot-Based Method of Testing and Adjusting for Publication Bias in Meta-Analysis,” Biometrics, 56, 455–463.
- Edwards, J. E., Oldman, A., Smith, L., Collins, S. L., Carol, D., Wiffen, P. J., McQuay, H. J., and Moore, R. A. (2000), “Single Dose Oral Aspirin for Acute Pain,” The Cochrane Database of Systematic Reviews, 2, CD002067.
- Egger, M., Davey Smith, G., Schneider, M., and Minder, C. (1997), “Bias in Meta-Analysis Detected by a Simple, Graphical Test,” British Medical Journal, 315, 629–634.
- Elfenbein, H. A., and Ambady, N. (2002), “On the Universality and Cultural Specificity of Emotion Recognition: A Meta-Analysis,” Psychological Bulletin, 128, 203–235.
- Harbord, R. M., Egger, M., and Sterne, J. (2006), “A Modified Test for Small-Study Effects in Meta-Analyses of Controlled Trials With Binary Endpoints,” Statistics in Medicine, 25, 3443–3457.
- Henmi, M., and Copas, J. (2010), “Confidence Intervals for Random Effects Meta-Analysis and Robustness to Publication Bias,” Statistics in Medicine, 29, 2969–2983.
- Higgins, J. P. T., and Green, S. (2011), Cochrane Handbook for Systematic Reviews of Interventions, Version 5.1.0. The Cochrane Collaboration. Available at http://handbook.cochrane.org/.
- Higgins, J. P. T., and Spiegelhalter, D. J. (2002), “Being Sceptical About Meta-Analyses: A Bayesian Perspective on Magnesium Trials in Myocardial Infarction,” International Journal of Epidemiology, 31, 96–104.
- Higgins, J. P. T., and Thompson, S. (2002), “Quantifying Heterogeneity in a Meta-Analyses,” Statistics in Medicine, 21, 1539–1558.
- Ioannidis, J. P. A. (2005), “Why Most Published Research Findings are False,” Chance, 18, 40–47.
- Moreno, S. G., Sutton, A. J., Ades, A. E., Stanley, T. D., Abrams, K. R., Peters, J. L., and Cooper, N. J. (2009), “Assessment of Regression-Based Methods to Adjust for Publication Bias Through a Comprehensive Simulation Study,” BMC Medical Research Methodology, 9, 2.
- Onion, R. (2008), “Reclaiming the Machine: An Introductory Look at Steampunk in Everyday Practice,” Neo-Victorian Studies, 1, 138–163.
- Paule, R., and Mandel, J. (1982), “Concensus Values and Weighting Factors,” Journal of Research of the National Bureau of Standards, 87, 377–385.
- Peters, J. L., Sutton, A. J., Jones, D. R., Abrams, K. R., Rushton, L., and Moreno, S. G. (2010), “Assessing Publication Bias in Meta-Analyses in the Presence of Between Study Heterogeneity,” Journal of the Royal Statistical Society, Series A, 173, 575–591.
- Rosenthal, R. (1979), “The File Drawer Problem and Tolerance for Null Results,” Psychological Bulletin, 86, 638–641.
- Rubin, D. B. (2005), “Causal Inference Using Potential Outcomes,” Journal of the American Statistical Association, 100, 322–331.
- Rücker, G., Schwarzer, G., Carpenter, J., Binder, H., and Schumacher, M. (2011), “Treatment-Effect Estimates Adjusted for Small Study Effects via a Limit Meta-Analysis,” Biostatistics, 12, 122–142.
- Sterne, J., and Egger, M. (2001), “Funnel Plots for Detecting Bias in Meta-Analysis: Guidelines on Choice of Axis,” Journal of Clinical Epidemiology, 54, 1046–1055.
- Sterne, J., Sutton, A., Ioannidis, J., Terrin, N., Jones, D. R., Lau, J., Carpenter, J., Rücker, G., Harbord, R. M., Schmid, C. H., Tetzlaff, J., Deeks, J. J., Peters, J., Macaskill, P., Schwarzer, G., Duval, S., Altman, D. G., Moher, D., Higgins, J. P. T. (2011), “Recommendations for Examining and Interpreting Funnel Plot Asymmetry in Meta-Analyses of Randomised Controlled Trials,” British Medical Journal, 343, d4002.
- Tanenbaum, J., Tanenbaum, K., and Wakkary, R. (2012), “Steampunk as Design Fiction,” in Proceedings of the SIGCHI Conference on Human Factors in Computing Systems. New York: ACM, pp. 1583–1592.
- Thompson, S., and Sharp, S. (1999), “Explaining Heterogeneity in Meta-Analysis: A Comparison of Methods,” Statistics in Medicine, 18, 2693–2708.
- Vansteelandt, S., and Joffe, M. (2014), “Structural Nested Models and g-Estimation: The Partially Realized Promise,” Statistical Science, 29, 707–731.
- Veroniki, A., Jackson, D., Viechtbauer, W., Bender, R., Bowden, J., Knapp, G., Kuss, O., Higgins, J. P. T., Langan, D., and Salanti, G. (2016), “Methods to Estimate the Between-Study Variance and Its Uncertainty in Meta-Analysis,” Research Synthesis Methods, 7, 55–79.
Appendix: R code for Section 4.3
Below we provide simple R code to verify that the estimating equation formulation of Egger regression in Section 4.3 is equivalent to fitting a weighted linear regression using the lm()function.
> ## G-estimation routine given Aspirin data vectors y,s
> G_est = function(a){
+ w = 1/s^2 ; Beta0 = a[1]
+ yBeta0 = y - Beta0*s ; MU = sum(w*yBeta0)/sum(w)
+ L = (sum(w*(yBeta0-MU)*(s - mean(s))))^2
+ }
> Beta0hat = optimize(G_est,c(-5,5))$min
> yBeta0hat = y - Beta0hat*s
> mu = sum(w*yBeta0hat)/sum(w)
> Beta0hat
[1] 2.112803
> mu
[1] 0.02519816
> ## standard Egger regression
> summary(lm(y~s,weights=1/s^2))$coef
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.0252 0.1884 0.134 0.894
s 2.1128 0.3122 6.767 5.81e-09 ***
> # Std. error estimation
> phi = summary(lm(y~s,weights=1/s^2))$sigma^2
> SE_MU = sqrt(phi/sum((1/s - mean(1/s))^2))
> SE_Beta0 = sqrt((SE_MU^2)*mean(1/s^2))
> SE_MU
[1] 0.1883619
> SE_Beta0
[1] 0.3122023

