Abstract
Capsule Atlantic Island Wrens are very closely related to mainland European populations.
Aims The first genetic screen of the North-east Atlantic island subspecies of (Winter) Wren Troglodytes troglodytes was performed to resolve their relationship to mainland Eurasian and Nearctic populations.
Methods The ND2 gene was sequenced from 15 wrens from Iceland, Faroe Islands, St Kilda, Outer Hebrides, Fair Isle and Shetland (2–3 individuals of each subspecies) and compared with Eurasian Wrens from mainland Britain and Europe, and Winter Wrens from North America.
Results All island subspecies were shown to originate from European rather than Nearctic founders. Genetic divergence from mainland British and European populations was small (0.1–0.3% uncorrected). The major European haplotype was present in some individuals from Shetland, Fair Isle and Faroes. Novel unique haplotypes were found in all individuals of St Kilda, Iceland and Hebridean Wrens, and in two individuals of Fair Isle Wren, contrasting with the high inferred levels of gene flow across Europe.
Conclusions Genetic data are consistent with a postglacial colonization of Atlantic islands from mainland Britain and Europe, possibly with continued gene flow due to migration of European birds. Tentatively, St Kilda Wren and Iceland Wren may be more closely related; most other subspecies are probably polyphyletic.
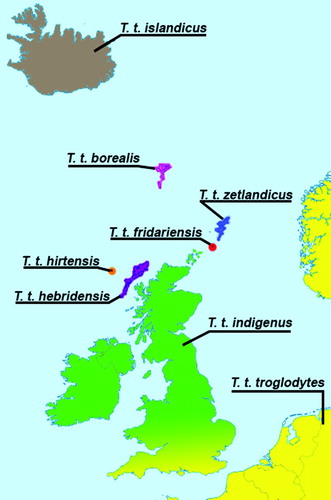
The Winter/Pacific/Eurasian Wren complex Troglodytes hiemalis/pacificus/troglodytes comprises a Holarctic grouping of insectivorous passerines with 41 recognized subspecies, and six distinct genetic clades currently classified as two or three species (Dickinson 2003, Drovetski et al. Citation2004, Toews & Irwin Citation2008, American Ornithologists' Union 2010). Six subspecies of the Eurasian Wren T. troglodytes inhabit oceanic islands in the North-east Atlantic, distinguishable on mean parameters of size, song, darkness and hue of plumage, and extent of barring on the upper and underparts (reviewed in McGowan et al. Citation2003, Armstrong Citation1955). These are the Shetland Wren T. t. zetlandicus, Fair Isle Wren T. t. fridariensis, Faroe Islands Wren T. t. borealis, Iceland Wren T. t. islandicus, Hebridean Wren T. t. hebridensis and St Kilda Wren T. t. hirtensis. In addition, a mainland British subspecies, T. t. indigenus, is often recognized, intergrading into nominate troglodytes in the south-east of Britain. Morphological and biometric summaries of these subspecies are included in Cramp et al. (Citation1988). Although all the island taxa are morphologically quite variable, there is a tendency to increased size c.f. nominate troglodytes, most obvious in Iceland Wrens. Plumage tones vary from generally colder brown/buff of hirtensis and fridariensis, to warmer (hebridensis) and darker (zetlandicus, borealis and islandicus). The different island races have mean differences in song structure and length – with a tendency for shorter, simpler songs than nominate birds (Cramp et al. Citation1988).
Although they are all (with the possible exception of indigenus) morphologically and vocally distinct from nominate troglodytes, no genetic data has been collected from the isolated island subspecies of the North-east Atlantic. It is likely that all subspecies colonized the islands within the last 12–22 000 years, during the current interglacial or since the Loch Lomond Stadial. While it can be assumed that colonization was from mainland Europe, it is not known whether all islands were colonized independently from the mainland, or whether island-hopping occurred. Furthermore it is possible that the St Kilda and Iceland subspecies in particular could have originated from Nearctic vagrants. Given these questions, a phylogeographic study of these island subspecies is warranted.
Some of the island subspecies exist in critically low population sizes and an understanding of their phylogeography and degree of genetic divergence is fundamental to any conservation attention they may receive. The Fair Isle Wren is of particular conservation concern: nesting only on sea cliffs of one island measuring 7.7 km2, its population has fluctuated between 10 and 50 singing males over the last 60 years (Williamson Citation1965, Pennington et al. Citation2004) and currently estimated at 29 singing males (Aspinall & Aspinall Citation2011, Musgrove et al. Citation2013), the Fair Isle Wren is highly susceptible to extinction by means of a stochastic event. The Shetland Wren has a population of 1500–3000 pairs and is found across all of Shetland with the exception of Fair Isle (Pennington et al. Citation2004, Forrester et al. Citation2007). Britain's other two subspecies are both found in the Outer Hebrides. The St Kilda Wren, as its name suggests, is confined to the archipelago of St Kilda. Williamson (Citation1958) calculated that there were 233 pairs distributed across the islands in 1957, but the population appears to be relatively stable, because a census in 2002 suggested that there were still around 230 pairs (Miles Citation2011). The Hebridean Wren occupies the rest of the Outer Hebrides including the Shiant Isles. The population is thought to be in the region of 5000–10 000, although no complete census has been made (Forrester et al. Citation2007, Musgrove et al. Citation2013). The final two subspecies are found on Iceland and the Faroe Islands, respectively. The population on the Faroe Islands is thought to be between 600–850 pairs (Bengtson Citation2001) and that on Iceland 3–5000 pairs (Hilmarsson Citation2011).
All of the island subspecies are thought to be either sedentary or very short-distance migrants. However, birds of the nominate subspecies are known to migrate south from northern continental Europe to over-winter in a less extreme climate (Parkin & Knox Citation2010). Birds identified on the basis of biometrics and/or plumage as likely nominate continental birds have been recorded migrating to or through the islands occupied by the Atlantic subspecies. The number of these birds is unknown, although counts of 30–40 have been made in locations such as Fair Isle and Sumburgh, Shetland, mostly during October (Shetland Bird Reports; Parkin & Knox Citation2010). While these autumn and winter records do not suggest that individuals may stay in Britain to breed the following year, there is a summer record of an apparent nominate Eurasian Wren on St Kilda in July 1984 (Murray Citation2002). It is therefore far from certain whether the Atlantic island subspecies are truly genetically isolated from mainland British and other European populations.
In this study, the mitochondrial NADH dehydrogenase 2 (ND2) gene was sequenced from all the Atlantic island subspecies and compared with ND2 sequences of wrens from elsewhere in Eurasia and the Nearctic. This extends on the work of Drovetski et al. (Citation2004), who also used the ND2 gene to assess the phylogenetic relationships of many other Winter Wren subspecies from Europe, Asia and North America (including indigenus, but none of the other island subspecies). The new data in the current study show that all the Atlantic subspecies had a Palearctic, not Nearctic, origin and provides preliminary evidence that the St Kilda and Iceland subspecies have experienced a period of isolation from mainland European birds.
METHODS
Sample selection
Toe pads of all Atlantic island subspecies were selected from verified specimens of certain identification and provenance held at National Museums Scotland. Fresh feathers dropped incidentally from locally breeding individuals or their known juveniles during routine ringing (banding) activities in Shetland were collected. In one case from Shetland, feathers were taken from a local juvenile found dead. A total of 15 samples were processed for this preliminary screen. Complete (>1 kb) ND2 sequence was obtained from 2–3 individuals of each island subspecies. A full list of the samples used for analysis can be found in .
Table 1. List of samples used for phylogenetic analysis.
DNA isolation and sequencing
Feather samples from fresh birds, and toepad samples from museum specimens, were used to isolate DNA, using the DNA Micro Kit (Qiagen, UK) with addition of dithiothreitol to 0.1 M concentration in the digestion mix and elution in 80 μl of Qiagen buffer AE. A polymerase chain reaction (PCR) fragment encompassing the entire coding region of ND2 was amplified from fresh samples using primers and conditions described in Drovetski et al. (Citation2004). To enable the successful amplification of the ND2 gene from fragmented DNA in museum specimens, primers were designed to amplify overlapping small fragments of the gene, each 150–250 bp in length (listed in ). Primers L5216 and H6313 are universal bird primers (Sorenson et al. Citation1999), while all TROG primers were designed against relatively conserved regions of T. troglodytes/hiemalis ND2 sequence collected from Genbank for use in this study. The fragments were designed to overlap to control for the possibility of redundant primer sequences introducing new single nucleotide polymorphisms (SNPs) in the specimen sequences analysed ().
Table 2. Primers used to sequence the ND2 gene in T. troglodytes subspecies.
PCR was performed using BIO-X-ACT Short thermostable DNA Polymerase (Bioline, UK). Each 50 µl PCR reaction contained 28.5 µl of ddH2O, 5 µl of 10x Optibuffer, 1 µl of 50 mM MgCl2 solution, 3 µl of dNTPs (2 mM each), 5 µl of forward and reverse primers (10 mM each), 2 units (0.5 µl) of DNA Polymerase and 2 µl of template DNA. The PCR thermal cycling programme was as follows: 3 minutes of template DNA denaturation at 95°C, then 35 cycles of denaturation at 95°C for 30 seconds, primer annealing at 55°C for 30 seconds and primer extension at 72°C for 30 seconds, followed by a final extension of 72°C for 10 minutes (Toews & Irwin Citation2008).
PCR products were separated by electrophoresis on a 1.5% agarose gel. The DNA from each gel fragment was then isolated using the QIAquick Gel Extraction Kit (Qiagen, UK) according to the manufacturer's protocols. Gel extracted PCR products were then diluted to 1 ng/µl/100 bp and sequenced by the Source BioScience LifeSciences (Cambridge) DNA sequencing service.
Data analysis
Using MEGA (Kumar et al. 1993), sequences were aligned using CLUSTAL (Higgins & Sharp 1992). Variable sites were located and unique haplotypes identified. Three phylogenies of the 13 haplotypes were constructed with the minimum evolution, neighbour joining and maximum parsimony methods, using evolutionary distances calculated with the Tamura–Nei model (Tamura & Nei 1993). Following Zhang & Gu (Citation1998), Gamma rate variation was set at α = 1.31. To examine the stability of clades, 1000 bootstrap replicates were used. Genetic variation was then assessed both for individuals within populations and between populations. A frequency distribution of these differences was then produced.
RESULTS
A total of 1030 nucleotides of the 1041 bp ND2 gene sequence could be unambiguously resolved from all 15 Atlantic island individuals sequenced. Sequences have been uploaded to the European Nucleotide database and their accession numbers are listed in . Added to this dataset were the ND2 gene sequences of 19 individuals from the NCBI Nucleotide database (http://www.ncbi.nlm.nih.gov/nuccore). Six of these individuals belong to the subspecies T. t. indigenus (accession numbers AY460295–300), 12 belonging to nominate T. t. troglodytes (accession numbers AY460301–311 and GQ369683) and a single sequence from Nearctic hiemalis (accession number AY460268) – total 34 ND2 sequences.
Using only the Eurasian samples (n = 33), a total of 16 variable sites were identified, which defined 12 unique haplotypes (). The inclusion of the T. hiemalis sequence increased the number of variable sites to 56. Of the 16 variable sites found in European birds, 12 involved transition mutations and 4 involved transversion mutations. Four of the haplotypes are shared by more than one individual, with one haplotype shared by 19 individuals across 5 subspecies. Nominate T. t. troglodytes showed exceptionally low genetic diversity, with one base change separating a single individual from the remaining 11, which all shared haplotype 1. In general, the Atlantic Island subspecies were genetically extremely close to nominate troglodytes, with no individual showing more than 3 bp difference (0.29% uncorrected) from the major haplotype 1. All three sequenced specimens of zetlandicus were genetically identical to this major haplotype, as were one individual each of fridariensis and borealis. All subspecies haplotypes were at least 31 bp divergent from Nearctic birds. Overall, it was concluded that all the isolated Atlantic Island subspecies are of European origin and that there has been no substantial genetic divergence since the islands were colonized.
Table 3. Polymorphic nucleotide sites defining the 12 ND2 haplotypes resolved from the 33 T. troglodytes sequences examined. Sequences are given in relation to haplotype 1, where a dot indicates identity. The subspecies that each haplotype was found in is listed.
The two specimens of St Kilda Wren (collected 88 years apart) shared a unique haplotype, 3 bp different from haplotype 1, which has not been observed in any other wren taxon. Similarly, both Iceland Wrens had a unique private haplotype, 2 bp different from major haplotype 1, though sharing the A → G SNP at position 174 shown by St Kilda Wrens. Each of the three Fair Isle Wrens had a different haplotype: one sharing major haplotype 1 with mainland European troglodytes as described above, and 2 with unique haplotypes, each 2 bp different from major haplotype 1. All 3 Hebridean Wrens had novel ND2 sequences – 2 with the same single 1 bp change (A → G at position 621 shared with 2 Fair Isle Wrens) and one with an additional 2 bp changes. Aside from the Faroes Wren with major haplotype 1 described above, the second individual had a further unique sequence with 2 bp changes not shown by any other wren taxon.
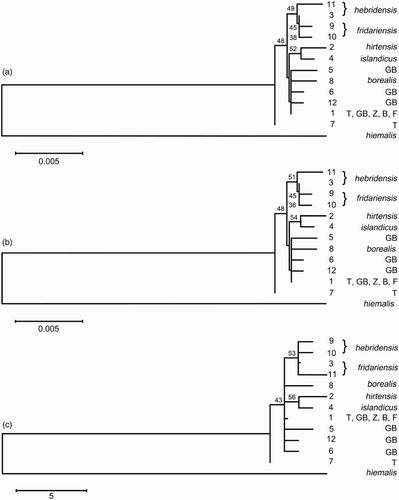
The potential phylogenetic relationships between the 12 haplotypes are shown in . The level of differentiation between T. troglodytes subspecies is very low, which precludes a very robust phylogeny based on the sequences obtained, all three analyses were consistent with St Kilda and Iceland subspecies being monophyletic, with T. t. hirtensis being the more divergent from the dominant continental haplotype. Paraphyly or monophyly of hebridensis was not well resolved. All other wren taxa appeared polyphyletic. The Fair Isle Wren's two unique haplotypes form a monophyletic clade with low support.
Figure 2. Minimum evolution (a) and neighbour joining (b) phylogenies of 13 Troglodytes ND2 haplotypes based on the Tamura–Nei distance with a gamma shape parameter of 1.31. Maximum parsimony (c) phylogeny shows most parsimonious tree with length = 58. Numbers at nodes denote bootstrap confidence indexes > 30%. Haplotype numbers (as per ) are given to the right of branch termini. T = nominate troglodytes, GB = birds from mainland Great Britain (troglodytes/indigenus), Z = zetlandicus, B = borealis and F = fridariensis.
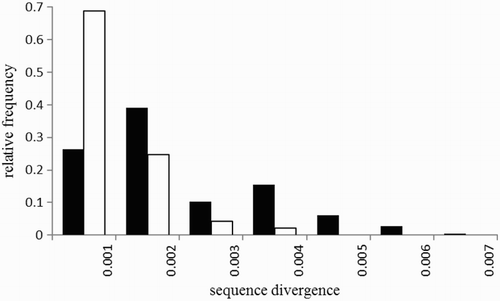
Levels of sequence divergence among wrens of the same subspecies ranged from 0 to 0.0032 (mean = 0.00047, 95% confidence interval = 0.0002–0.0008), while divergence between types ranged from 0 to 0.0062 (mean = 0.00174, 95% confidence interval = 0.0015–0.0020). shows the frequency distributions of sequence divergence within and between wren taxa.
DISCUSSION
Drovetski et al. (Citation2004) noted that vicariance was the key to the development of the six clades of Winter/Pacific/Eurasian Wren. However, the study noted the absence of individuals from the island races of the North-east Atlantic and postulated that it was plausible that birds from Europe or from the eastern Nearctic could have originally colonized these islands. The current study suggests that all the North-east Atlantic subspecies are much more closely related to nominate T. t. troglodytes than the Nearctic hiemalis outgroup. This was to be expected given the islands' proximity to continental Europe and the lack of a Winter Wren population in Greenland, which would have been the most likely route for dispersal from North America.
Nucleotide diversity across all European Wrens was very low, however birds from the insular races including mainland Britain showed much greater diversity (mean base changes per site, d = 0.00215) than continental birds (d = 0.00016). The homogeneity displayed in nominate T. t. troglodytes, where a single base change separated all 12 individuals, may be explained by migration. Northerly populations of T. t. troglodytes migrate south from higher latitudes to overwinter in a warmer climate. This annual mixing of the population can lead to genetic homogeneity, assuming at least some long-distance dispersal of birds that remain to breed in a new geographic area (Oomen et al. Citation2011). This would explain how individuals from France, Germany, Russia and Switzerland all shared an identical haplotype.
Migration may also be a key factor in understanding why many of the insular races are apparently not monophyletic. With reports of large numbers of nominate birds migrating through Shetland and Fair Isle (Parkin & Knox Citation2010), it is possible that the occasional continental individual lingers on the islands to breed. A single breeding migrant per generation can start to cause genetic homogeneity between populations, thus diluting any phylogenetic signal as the dominant haplotype is mixed into the gene pools on each island (Mills & Allendorf Citation1996). This may explain why all individuals sampled from Shetland shared the dominant nominate haplotype.
Iceland and St Kilda are very isolated geographically, and do not fall into the path of usual migration route of Scandinavian birds moving south. Given the observation that these taxa are sedentary, there may therefore be little or no opportunity for gene flow. Both races have their own haplotype that is endemic to their respective ranges (). The haplotypes of these two races share a polymorphism of a single nucleotide that is not shared with any of the other haplotypes identified. Sample sizes are small and further sampling is required to test a hypothesis of an ancestral link between the two populations.
Another interesting race is the Hebridean Wren, because the three samples were defined by two haplotypes that were distinct to the subspecies and phylogenetic analysis showed them to be either paraphyletic or monophyletic depending on the tree construction method used. The two individuals collected on Hyskeir share a haplotype, while the bird from South Uist, Outer Hebrides, is more divergent. The two haplotypes belonging to T. t. hebridensis share a nucleotide variant compared to haplotype 1, yet the South Uist bird has subsequently developed a further two base changes. The South Uist bird was also found to be one of the most divergent individuals, with six base differences between its haplotype and the St Kilda haplotype, as shown in . This is unexpected given the proximity of St Kilda to the Outer Hebrides.
Forrester et al. (Citation2007) stated that Wren specimens from the Isle of Skye were found to be of the race T. t. indigenus and as such the range of the Hebridean Wren was thought restricted to the Outer Hebrides. Hyskeir is located 14 km to the west of the Inner Hebridean islands of Canna and Rum, 40 km to the east of the Outer Hebridean Island of Barra. While Hyskeir birds are currently attributed to subspecies hebridensis, further sampling of birds both from here and from the major Outer Hebridean islands could determine whether Hyskeir birds potentially represent a genetically isolated population.
The three individuals from Fair Isle yielded surprisingly diverse haplotypes given the extremely small population on the island. Two Fair Isle haplotypes tentatively form a monophyletic clade, while one individual (F1) had the dominant haplotype 1. Haplotype 1 was also present in Faroese birds. This may indicate that migratory continental birds are occasionally finding their way to the Faroe Islands, despite it being geographically more isolated than the British Isles. Alternatively, haplotype 1 may represent an ancestral genotype which has remained in the population since the initial colonization of the islands of the North-east Atlantic.
Given the clear morphological differences between the insular races of Eurasian Wren, the level of genetic homogeneity was unexpectedly high. Although our sample sizes are small, the preliminary data suggest a pattern similar to that observed for Pacific Wrens T. pacificus from Kodiak, the Pribilof and Aleutian Islands (Pruett & Winker Citation2008, Toews & Irwin Citation2012). Several of the island subspecies of Pacific Wren are morphologically and vocally distinct from each other and from mainland populations, but levels of cytochrome b genetic divergence are very low, with novel island haplotypes suggestive of a period of glacial isolation (Pruett & Winker Citation2008). It can be suggested that natural selection has acted rapidly to drive significant morphological change of isolated populations of these wren taxa in what are sometimes marginal habitats.
Williamson (Citation1958) suggested that the North-east Atlantic races could be split into three groups based on phenotypic characters. The groups were the greyer St Kilda and Fair Isle races, the largest and darkest northern races from Iceland and the Faroes, and a final group with intermediate traits between the northern races and birds from mainland Britain consisting of T. t. hebridensis and T. t. zetlandicus. The preliminary data described here do not support this arrangement. The similarity in colouration between T. t. hirtensis and T. t. fridariensis may be due to similar ecological traits. Both races share a preference for coastal habitat and forage on rocky shorelines. Meanwhile the extreme body sizes found in the northern-most races may be an example of Bergmann's rule (Bergmann Citation1847), which is often associated with larger body sizes being observed at higher latitudes. Further sequencing would need to be performed to determine whether fixation of a unique haplotype has occurred in the most remote islands, Iceland and St Kilda, which would indicate that these populations are isolated both geographically and reproductively (Frankham Citation1997).
As Drovetski et al. (Citation2004) noted that there is very low nucleotide diversity within the nominate continental European race of T. troglodytes. The relatively small level of genetic variation within and between the insular subspecies could therefore nevertheless be considered significant. Subspecies that are found to be monophyletic with diagnostic morphometric or other phenotypic features and one or more unique haplotypes, potentially St Kilda, Hebridean and Iceland Wrens, could be considered as evolutionary species (Helbig et al. Citation2002) if further sampling of more individuals showed that they were reciprocally monophyletic with respect to each other and other wren taxa. An analysis of multi-locus nuclear markers may also be informative.
Further research would be desirable to increase the sample sizes from each subspecies, and to use the more genetically variable control region of mtDNA to provide greater resolution of phylogenetic signal from these closely related taxa. This was attempted during this study however ‘universal’ avian control region primers did not work on the wren samples. Furthermore, the subspecific identification of wrens from multiple Scottish and Irish Islands remains uncertain, and should be resolved, both to provide information on phylogeography of the species and because there may be unrecognized genetically differentiated populations that merit subspecific status.
ACKNOWLEDGEMENTS
We are grateful to Roger Riddington and Fair Isle Bird Observatory staff for providing samples from Shetland Wren and Fair Isle Wren for this study.
REFERENCES
- American Ornithologists' Union. 2010. Forty-second supplement to the American Ornithologists' Union Check-list of North American Birds. Auk 127: 726–744.
- Armstrong, E.A. 1955. The Wren (New Naturalist Monograph). Collins, London.
- Aspinall, S. & Aspinall, R. 2011. The Fair Isle Wren: population and territory occupancy. Br. Birds 104: 312–324.
- Bengtson, S. 2001. Breeding distribution and numbers of wren (Troglodytes troglodytes) in the Faroe Islands. Fróðskaparrit 49: 127–139.
- Bergmann, C. 1847. Über die Verhältnisse der Wärmeökonomie der hiere zu ihrer Grösse. Göttinger Studien, 3: 595–708.
- Cramp, S., Brooks, D.J., Dunn, E., Gillmor, R., Hall-Craggs, J., Hollon, P.A.D., Nicholson, E.M., Ogilivie, M.A., Roselaar, C.S., Sellar, P.J., Simmons, K.E.L., Voous, K.H., Wallace, D.I.M. & Wilson, M.G. 1988. Birds of Europe, te Middle East and North Africa: The Birds of the Western Palearctic. Oxford University Press, Oxford.
- Dickinson, E.D. (ed.) 2003. The Howard and Moore Complete Checklist of the Birds of the World, 3rd edn. Christopher Helm, London.
- Drovetski, S.V., Zink, R.M., Rohwer, S., Fadeev, I.V., Nesterov, E.V., Karagodin, I., Koblik, E.A. & Red'kin, Y.A. 2004. Complex biogeographic history of a Holarctic passerine. Proc. R. Soc. B. 271: 545–551. doi: 10.1098/rspb.2003.2638
- Forrester, R., Andrews, I., McInerny, C., Murray, R., McGowan, R., Zonfrillo, B., Betts, M., Jardine, D. & Grundy, D. 2007. The Birds of Scotland. Scottish Ornithologists’ Club, Edinburgh.
- Frankham, R. 1997. Do island populations have less genetic variation than mainland populations? Heredity 78: 311–327. doi: 10.1038/hdy.1997.46
- Helbig, A., Knox, A., Parkin, D. T., Sangster, G. & Collinson, M. 2002. Guidelines for assigning species rank. Ibis 144: 518–525. doi: 10.1046/j.1474-919X.2002.00091.x
- Higgins, D.G. & Sharp, P.M. 1992. CLUSTAL V: Improved software for multiple sequence alignment. Comput. Appl. Biosci. 8: 189–191.
- Hilmarsson, J.O. 2011. Icelandic Bird Guide. Forlagið, Reykjavik.
- Kumar, S., Tamura, K. & Nei, M. 1993. MEGA: Molecular Evolutionary Genetics Analysis. Pennsylvania State University, University Park, PA.
- McGowan, R.Y., Clugston, D.L. & Forrester, R.W. 2003. Scotland's endemic subspecies. Scot. Birds 24: 18–35.
- Miles, W. 2011. The appearance and status of the St Kilda Wren. Br. Birds 104: 325–328.
- Mills, L.S. & Allendorf, F.W. 1996. The one-migrant-per-generation rule in conservation and management. Conserv. Biol. 10: 1509–1518. doi: 10.1046/j.1523-1739.1996.10061509.x
- Murray, S. 2002. Birds of St Kilda. Scot. Birds 23: suppl. 1–64.
- Musgrove, A., Aebischer, N., Eaton, M., Hearn, R., Newson, S., Noble, D., Parsons, M., Risely, K. & Stroud, D. 2013. Population estimates of birds in Great Britain and the United Kingdom. Br. Birds 106: 64–100.
- Oomen, R.A., Reudink, M.W., Nocera, J.J., Somers, C.M., Green, M.C. & Kyle, C.J. 2011. Mitochondrial evidence for panmixia despite perceived barriers to gene flow in a widely distributed waterbird. J. Hered. 102: 584–592. doi: 10.1093/jhered/esr055
- Parkin, D.T. & Knox, A.G. 2010. Status of Birds in Britain and Ireland. A & C Black Ltd, London.
- Pennington, M., Ellis, P., Harvey, P., Heubeck, M., Okill, D., Osborn, K. & Riddington, R. 2004. The Birds of Shetland. Christopher Helm, London.
- Pruett, C.L. & Winker, K. 2008. Evidence for cryptic northern refugia among high- and temperate-latitude species in Beringia. Climate Change 86: 23–27. doi: 10.1007/s10584-007-9332-6
- Sorenson, M.D., Ast, J.C., Dimcheff, D.E., Yuri, T. & Mindell, D.P. 1999. Primers for a PCR-based approach to mitochondrial genome sequencing in birds and other vertebrates. Molec. Phylogenet. Evol. 12: 105–114. doi: 10.1006/mpev.1998.0602
- Tamura, K. & Nei, M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10: 512–526.
- Toews, D.P.L. & Irwin, D.E. 2008. Cryptic speciation in a Holarctic passerine revealed by genetic and bioacoustic analyses. Molec. Ecol. 17: 2691–2705. doi: 10.1111/j.1365-294X.2008.03769.x
- Toews, D.P.L. & Irwin, D.E. 2012. Pacific Wren (Troglodytes pacificus). In Poole, A. (ed.) The Birds of North America Online. Cornell Lab of Ornithology, Ithaca. Retrieved from http://bna.birds.cornell.edu/bna/species/720
- Williamson, K. 1958. Population and breeding environment of the St Kilda and Fair Isle Wrens. Br. Birds 51: 369–393.
- Williamson, K. 1965. Fair Isle and its Birds. Oliver & Boyd, Edinburgh.
- Zhang, J. & Gu, X. 1998. Correlation between the substitution rate and rate variation among sites in protein evolution. Genetics 149: 1615–1625.