ABSTRACT
1. Bromodomain-containing protein 2 (BRD2) is an important member of the BET protein family, which can specifically bind histone acetylated lysine to participate in gene transcriptional regulation, chromatin remodelling, cell proliferation and apoptosis. The following investigation of cellular proteins interacting with chBRD2 will be helpful in understanding the new functions of chBRD2 and the mechanism of NDV replication.
2. The recombinant eukaryotic expression vector pEGFP-chBRD2 and empty vector pEGFP-C1 were transfected into DF-1 cells to overexpress GFP-chBRD2 and GFP, respectively. GO annotation, KEGG pathway, and protein-protein interaction network were used to analyse the cellular proteins interacting with chBRD2. In addition, one targeted protein was selected to verify its interaction with chBRD2 using fluorescent co-localisation and Co-IP.
3. A total of 225 cellular proteins were identified that potentially interact with chBRD2. GO analysis showed that these play key roles in gene transcriptional regulation, cell cycle and development, immunity and viral infection. Further KEGG pathway analysis showed that these proteins were mainly involved in genetic information processing, immune system, cellular processes and translation. In addition, one targeted cellular protein chicken matrin 3 (chMATR3) was also identified as chBRD2 complex using both fluorescence co-localisation and Co-IP analysis.
4. This study presents the interactome data of chBRD2 protein in DF-1 cells, which provides new information to understand the functions of chBRD2 and new targets for further investigating the replication and pathogenesis of NDV.
Introduction
The Bromodomain and Extra-Terminal Domain (BET) family proteins can regulate gene transcription by modulating chromatin opening, facilitating transcription factor recruitment to target gene promoter and enhancer sites, and promoting activation of paused RNA polymerase II (Pol II) transcriptional machinery for gene transcription elongation (Chiang Citation2009; Tomohiko et al. Citation2014). Bromodomain-containing protein 2 (BRD2) is an important member of the BET family (comprising BRD2, BRD3, BRD4, and testis-specific BRDt), which contains two tandem bromodomain functional domains (BD1 and BD2), a nuclear localisation signal domain (NLS) and an extra-terminal (ET) domain (Florence and Faller Citation2001). The BD domain was first found in drosophila brahma protein, which is a highly conserved domain composed of about 110 amino acid residues and can specifically recognise and bind the acetylated lysine residues in histone, while the ET domain is a conservative domain composed of about 80 amino acids, which regulates gene transcription by recruiting specific effector proteins (Zeng and Zhou Citation2002; Rahman et al. Citation2011). The high selectivity of BRD2, when binding to acetylated lysine 12 of histone H4, may provide a mechanistic basis for the hypothesised ‘histone code’ of acetylation, phosphorylation, methylation, and other post-translational modifications (PTMs) of nucleosomal histones that contribute to transcription regulation (Kanno et al. Citation2004). Hence, BRD2 can recruit coactivators (histone acetylases) to upregulate transcription of the target gene or corepressors (histone deacetylases) to downregulate transcription of the target gene in a versatile fashion (Sinha et al. Citation2005). It has been shown that BRD2 protein binds to DNA double-strand breaks, regulates DNA damage and repair and interacts with the limiting replication initiation protein TICRR to regulate replication initiation (Gursoyyuzugullu et al. Citation2017; Sansam et al. Citation2018). Meanwhile, BRD2 has the function of a cytoskeleton protein and transcriptional activation to promote the recruitment of E2F-1 and chromatin remodelling activity in the process of E2F-1 regulating the progress of cell cycle (Peng et al. Citation2007).
Three types of cellular proteins interact with human BRD2 (huBRD2) protein, including chromatin-related proteins, transcriptional activation-related proteins and cell cycle regulatory-related proteins (Denis et al. Citation2006). This has increased the understanding of the functions of BRD2. However, considering the low homology (77.5%) between huBRD2 and chicken BRD2 (chBRD2), the kind of cellular proteins interacting with chBRD2 and whether these cellular proteins are similar to that of huBRD2 remains unknown. In addition, several studies have reported an interaction of viral proteins with host BRD2 to promote virus replication (Viejoborbolla et al. Citation2003, Citation2005; Gallay et al. Citation2019). As for chicken BRD2 (chBRD2), a recent study has shown that Newcastle disease virus (NDV) matrix protein interacts with chBRD2 to enhance virus replication by promoting viral RNA synthesis and transcription (Duan et al. Citation2020), but more efforts are needed to determine how this interaction might influence NDV replication. Therefore, in this study, the recombinant eukaryotic expression vector pEGFP-chBRD2 and empty vector pEGFP-C1 were transfected into DF-1 cells to overexpress GFP-chBRD2 and GFP, respectively. Then the co-immunoprecipitation (Co-IP) was combined with liquid chromatography-tandem mass spectrometry (LC-MS) and used to screen and identify those cellular proteins interacting with chBRD2. These results provide valuable information in understanding the new functions of chBRD2 and investigating the replication and pathogenesis of NDV.
Materials and methods
Cells and antibodies
Chicken embryo fibroblast cells (DF-1) were purchased from the Cell Resource Center of Shanghai Institutes for Biological Sciences of the Chinese Academy of Sciences. Mouse anti-GFP monoclonal antibody (66002) and mouse anti-Myc monoclonal antibody (66003) and HRP-conjugated Affinipure Goat Anti-Mouse IgG (H + L) (SA00001) were purchased from Proteintech Company (Wuhan, China).
Plasmid construction
All enzymes used for cloning procedures were purchased from Thermo Scientific (USA). The plasmids pEGFP-C1 and pCMV-Myc were purchased from Clontech (USA), Total RNA was extracted from DF-1 cells by TRIzol and reverse transcribed. The open reading frame (ORF) of the chBRD2 (XM_015294960) and chicken matrin 3 (chMATR3) gene (NM_204147) were amplified by specific primers and then used to construct the recombinant expression plasmids pEGFP-chBRD2 and pCMV-Myc-chMATR3 (). Primers used for the construction of the above recombinant expression plasmids are shown in . All of the constructed recombinant expression plasmids were confirmed by PCR, restriction digestion and DNA sequencing.
Table 1. Primers used for the construction of recombinant expression plasmids
chBRD2 gene (GenBank accession: XM_015294960); chMATR3 gene (GenBank accession: NM_204147); Primers were designed by Primer Premier 5.0. pEGFP-chBRD2 and pCMV-Myc-chMATR3 were used to amplify the ORF of chBRD2 gene and chMATR3 gene; Underline and bold indicate restriction site.
Cell culture, transfection and fluorescence microscopy
The DF-1 cells were cultured in Dulbecco’s modified Eagle medium (DMEM; Gibco, USA) containing 15% foetal bovine serum (FBS; Gibco, USA) at 37°C under an atmosphere of 5% CO2. For the transfection experiments, 5 × 105 DF-1 cells were grown to 80% confluence in six-well plates, and then pEGFP-chBRD2 and pEGFP-C1 (3 μg of each plasmid) were respectively transfected into DF-1 cells with lipofectamine 3000 (Invitrogen, USA) according to the manufacturer’s instructions. Twenty-four hours after transfection, cells expressing the recombinant proteins GFP-chBRD2 and GFP were rinsed with phosphate-buffered saline (PBS), fixed with precooled 4% paraformaldehyde for 20 min at room temperature, permeabilised with 0.25% Triton X-100 for 5 min, and then counterstained with DAPI (Sigma, USA) to detect nuclei. Fluorescent images were obtained under an ortho fluorescence microscope and then analysed and merged using Adobe Photoshop CS7 software.
Co-IP assays
For Co-IP assays, 5 × 105 DF-1 cells cultured in six-well plates were transfected with the plasmids expressing GFP-chBRD2 and GFP-C1, respectively. At 36 h post-transfection, cells were washed three times with PBS and then lysed with immunoprecipitation lysis buffer (Invitrogen, USA). The supernatants were collected after centrifugation and then the chBRD2 and interacting proteins in DF-1 cells were enriched using the Pierce™ Classic Magnetic IP/Co-IP Kit 88 804 (Invitrogen, USA), according to the manufacturer’s instructions. The partial immune-precipitates were detected by Western blotting using anti-GFP antibody and the rest were stored at −80°C before LC-MS.
Protein identification and bioinformatics analysis
The Co-IP experiment was carried out using the Co-IP Kit (Pierce, USA). The final eluate of Co-IP (20 μl) was obtained after washing five times, and then used for LC-MS analysis. In addition, all proteins present in the negative controls were excluded and only those appearing at least twice in the triplicate analyses were reserved to generate interaction data. The LC-MS analysis was conducted by Wuhan GeneCreate Biological Engineering Co, Ltd. (Wuhan, China). ekspertTMnanoLC and the AB Sciex TripleTOF 5600-plus (USA) was used for MS data generation. Functional annotation and classification of all identified proteins were determined by using the Blast2GO program against the UniProt database (https://www.uniprot.org/). KEGG Pathway analyses were extracted using the search pathway tool (http://www.genom e.jp/kegg/mappe r.html). The protein-protein interaction (PPI) network and differential Venn diagrams were drawn by online software STRING Version 11.0 (https://string-db.org/cgi/input?sessionId=beWNfFR7fSbZ&input_page_show_search=on) and draw Venn diagrams (http://bioinformatics.psb.ugent.be/webtools/Venn/), respectively.
Verification of the protein-protein interaction
Based on the above results, the screened cellular protein chMATR3 was selected to verify its interaction with chBRD2, using fluorescence co-localisation and Co-IP assays. For the fluorescence co-localisation assay, 5 × 105 DF-1 cells were grown to 70% confluence in six-well plates, and then a total of 3 μg of the combined plasmids (pEGFP-chBRD2 and pCMV-Myc-chMATR3 or pEGFP-chBRD2 and pCMV-Myc or pEGFP-C1 and pCMV-Myc-chMATR3) were transfected into DF-1 cells (1.5 μg for each plasmid) using lipofectamine 3000, according to the manufacturer’s instructions. Thirty-six hours after transfection, cells were rinsed with PBS, fixed with precooled 4% paraformaldehyde for 20 min at room temperature, permeabilised with 0.25% Triton X-100 for 5 min, blocked with BSA for 1 h, and then incubated with mouse anti-Myc for 1 h. After three washes with PBS, cells were incubated with Alexa Fluor 488-conjugated goat anti-mouse IgG (H + L) for 1 h. Cells were counterstained with DAPI to detect nuclei. Fluorescent images were obtained under an inverted fluorescence microscope, and then analysed and merged using Adobe Photoshop CS7 software. For the Co-IP assay, the combined plasmids were transfected into cells and Co-IP assay was performed as described above to confirm the interaction between chBRD2 and chMATR3.
Results
Intracellular expression and subcellular localisation of GFP-chBRD2 and GFP
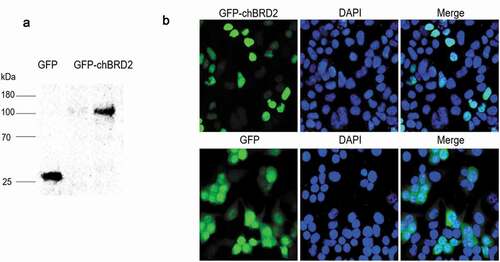
In order to investigate whether the GFP-chBRD2 recombinant and GFP proteins were expressed in DF-1 cells, the total proteins were extracted from the plasmid-transfected cells and then detected by Western blot analysis. The results showed that, after transfection of pEGFP-chBRD2 and pEGFP-C1 into DF-1 cells, protein bands of about 114 kDa and 28 kDa were observed (chBRD2: 85.66 kDa, GFP: 28 kDa), which were consistent with the predicted size ()). In addition, the results of fluorescence revealed that the recombinant protein GFP-chBRD2 was principally localised in the nucleus, while the GFP tag protein was found both in the nucleus and the cytoplasm ()).
Identification of cellular proteins interacting with chBRD2 protein
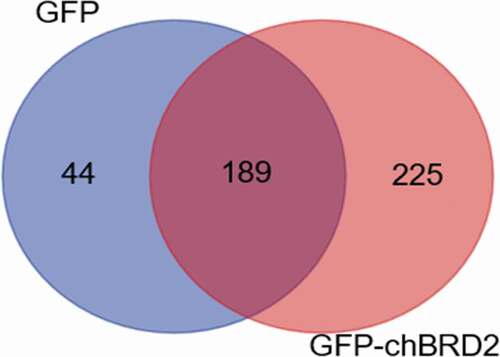
Protein identification showed that the total number of spectra in the negative control group (GFP) and the experimental group (GFP-chBRD2) were 15138 and 16450, respectively, and the resolved secondary spectrum numbers were 2959 and 3541, respectively (). A total of 233 proteins interacted with GFP and 414 with GFP-chBRD2 (Supplementary Table S1 and S2; ). However, only 225 cellular proteins (Supplementary Table S3) were found to interact with GFP-chBRD2 using Venn analysis ().
Table 2. Statistical table of protein identification information
Figure 2. The distribution of differentially expressed proteins during GFP-chBRD2 or GFP infection at 36 h. The differential Venn maps of proteins interacting with GFP-chBRD2 or GFP were completed by the online programme. (http://bioinformatics.psb.ugent.be/webtools/Venn/)
The results of MS identification were provided by Wuhan GeneCreate Biological Engineering Co. Ltd. (Wuhan, China). there were 253 proteins interacting with GFP and 428 proteins interacting with GFP-chBRD2.
Functional analysis of chBRD2 interacting proteins
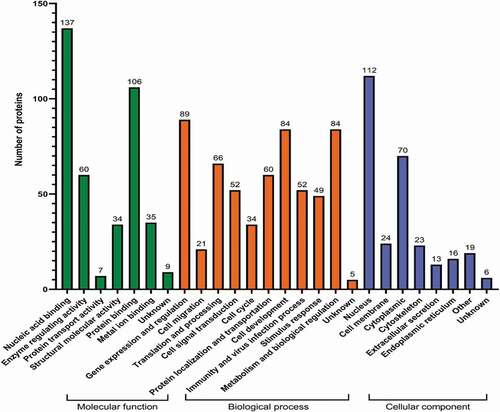
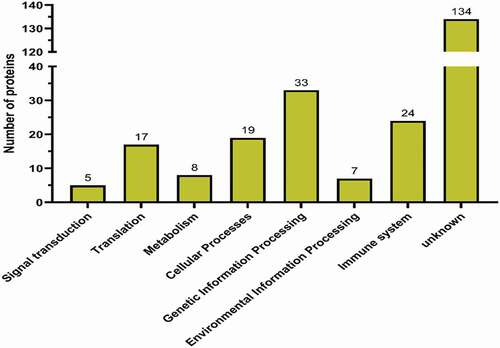
According to the Uniprot database, the proteins interacting with chBRD2 were annotated and classified. Gene ontology enrichment analysis showed that the most significantly enriched molecular functions of chBRD2 involved nucleic acid binding, protein binding and enzyme activity regulation. As for biological process enrichment, this was regulated by gene expression, cell development, biological regulation and metabolism, while, for the cellular components, most of the proteins were located in the nucleus, followed by the cytoplasm (). To obtain more information about the possible biological pathways involved in chBRD2, KEGG pathway enrichment analysis was included. In addition to ‘unknown’ proteins, cellular proteins interacting with chBRD2 were mainly involved in genetic information processing, immune system, cellular processes and translation ().
Figure 4. KEGG pathway analysis of differentially expressed proteins (http://www.genom e.jp/kegg/mappe r.html)
Network relationship of chBRD2 interacting proteins
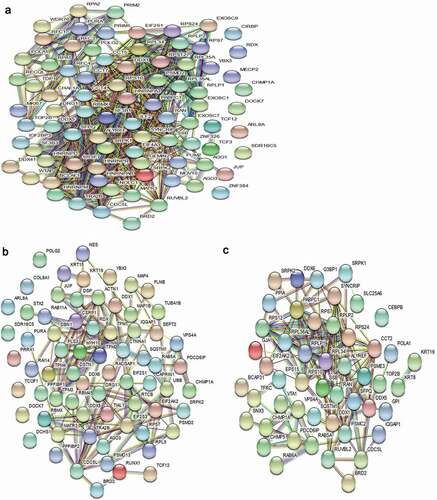
To investigate the information of the chBRD2-interaction network, a map was constructed by introducing proteins interacting with chBRD2 through the online software STRING (PPI). As 34 proteins contained no relevant information, the other 191 proteins were analysed. In order to display the information of PPI more intuitively, the results were imported into Cytoscape software for optimisation. The results showed that 179 out of 191 proteins had complex interactions using the PPI diagram (). Meanwhile, three types of network were constructed, including gene expression and regulation ()), cell cycle and cell development ()) and immunity and virus infection ()).
Figure 5. The PPI network diagram of proteins interacting with chBRD2. The colour, from red to yellow, indicates the number of proteins that interact with chBRD2 and a black line indicates that the interaction between the two proteins was predicted (https://string-db.org/cgi/input?sessionId=beWNfFR7fSbZ&input_page_show_search=on)
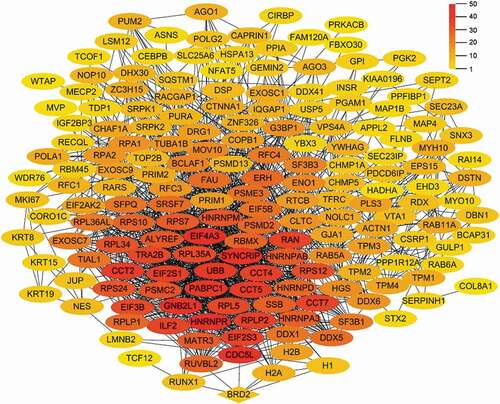
Figure 6. The PPI network diagram of proteins interacting with chBRD2 based on biological function. (a) The PPI network diagram was based on gene expression and regulation. (b) The PPI network diagram was based on cell cycle and cell development. (c) (https://string-db.org/cgi/input?sessionId=beWNfFR7fSbZ&input_page_show_search=on). The red line indicates the gene fusions evidence; the blue line gene co-occurrence evidence; the light blue line database evidence; the purple line experimental evidence, the light green text mining evidence; the green line neighbourhood evidence; the black line co-expression evidence and the light purple protein homology evidence
Characterisation of interaction between chBRD2 and chMATR3
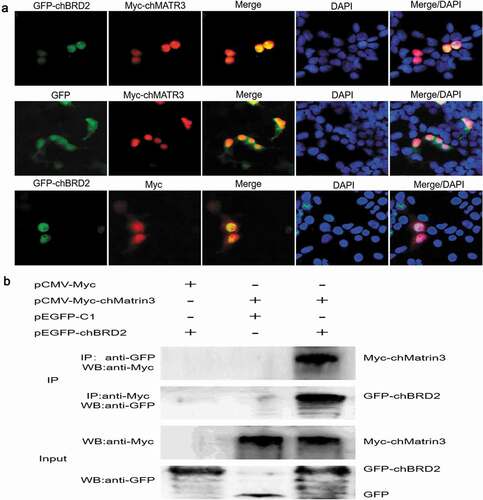
To further validate the Co-IP and LC-MS results, fluorescence co-localisation and Co-IP assay were used to examine the interaction between chBRD2 and chMATR3. The fluorescent co-localisation results revealed that chBRD2 and chMATR3 were co-localised in the nucleus ()). The Co-IP results showed that the fusion protein GFP-chBRD2, but not GFP tag, could be immunoprecipitated with Myc-chMATR3 when using an anti-Myc antibody, and the inverted experiment obtained the same result ()).
Figure 7. Identification of interaction between chBRD2 and chMATR3 by fluorescence co-localisation and Co-IP assays. (a) Co-localisation of chBRD2 and chMATR3 in plasmids co-transfected DF-1 cells. Original magnification was 1 × 200. (b) Co-IP assay was used to verify the interaction between chBRD2 and chMATR3
Discussion
The BET proteins play crucial roles in regulating gene transcription through epigenetic interactions between bromodomain and acetylated histones during cellular proliferation and differentiation processes. In the process of virus infection and integration of the virus genome into the host genome, it is equally important that BET protein mediates virus infection and replication through the interaction between ET domain and viral protein (Taniguchi Citation2016). As an important member of the BET proteins, BRD2 is known to participate in various cellular and biological processes, including chromatin remodelling (Peng et al. Citation2007), cell-cycle progression (Sinha et al. Citation2005), adipocyte differentiation (Zang et al. Citation2013), inflammatory responses (Belkina and Denis Citation2012) and virus replication (Viejoborbolla et al. Citation2005). In a previous study, chBRD2 from the cDNA library of DF-1 cells was screened to interact with NDV matrix protein by the yeast-two hybrid system (Duan et al. Citation2018). In addition, it was found that the NDV matrix protein interacted with the ET domain of chBRD2 and reduced chBRD2 expression to enhance virus replication by promoting viral RNA synthesis and transcription in DF-1 cells (Duan et al. Citation2020). To better understand the new functions of chBRD2 and investigate how NDV matrix protein-chBRD2 interaction may influence NDV replication, the current study screened and analysed the cellular proteins interacting with chBRD2 in DF-1 cells. Compared with a previous study reported by Denis et al. (Citation2006), cellular proteins interacting with huBRD2 were mainly chromatin, transcriptional activation- and cell cycle-related proteins. However, the screened cellular proteins interacting with chBRD2 were mainly related to gene transcriptional regulation, cell cycle and development, immunity and viral infection. Thus, these results provided useful information in understanding the new functions of chBRD2.
chBRD2 is involved in gene expression regulation
It is generally believed that histone acetylation leads to the loosening of chromatin structure and increases the accessibility of nucleosome DNA to transcription factors, thus activating gene transcription (Pham et al. Citation2018). It has been reported that BRD2 can bind acetylated histone H4 to recruit transcription factors, co-activators and co-inhibitors to regulate transcription (Umehara et al. Citation2010). For instance, BRD2 recruits HAT, HDAC, transcription-related proteins and chromatin remodelling complexes to participate in epigenetic modification, allowing different proteins to play their roles at appropriate periods (Leroy et al. Citation2008). A previous study has shown that the bromodomains and C-terminal domain of BRD2 are equally important for transcription and splicing regulation, which correlates with the role of these domains in BRD2 binding to chromatin (HnilicovÁ et al. Citation2013). Denis et al. (Citation2006) found that several cellular proteins, including histone H2A, H2B, H3 and H4, histone deacetylase 11 (HDAC11), chromatin assembly factor 1 (CAF1) and SWI/SNF-related chromatin regulator p270, that interact with huBRD2 are chromatin-related proteins. These may participate in chromatin remodelling and gene expression regulation. In the current study, certain chromatin-related proteins, such as histone H2A and H2B, CAF1, were found to interact with chBRD2, which was consistent with a previous study.
In addition, some other cellular proteins involved in gene expression regulation were identified. For example, CDC5L, part of the Prp19 complex that participates in pre-mRNA splicing and DNA damage repair (Sihn et al. Citation2007), and RUVBL2, together with TIP60, regulated transcription via chromatin remodelling by participating in diverse multi-protein complexes (Nishitsuji et al. Citation2018; Idauden et al. Citation2020). Studies have shown that there are at least 20 members of the heterogeneous nuclear ribonucleoprotein (HNRNP) family identified in mammals (Han et al. Citation2010), which perform a variety of functions related to transcription, splicing, RNA stability and degradation (Lu et al. Citation2006; Gittings et al. Citation2019). Interestingly, HNRNPAB, HNRNPD, HNRNPR, HNRNPM, HNRNPR, and HNRNPQ were screened for the ability to possibly form interaction complexes with chBRD2. Moreover, microtubule-associated protein, nuclear, nucleolar protein, replication, RNA-binding protein, splicing and transcription factors were found in the chBRD2 complex. More studies are needed to verify such interactions and investigate the role of these in gene expression regulation.
chBRD2 in cell cycle and differentiation
The study of the cell cycle controlled by the transcription of some key genes involves two aspects, (i) direct recruitment of sequence-specific DNA-binding transcriptional activators, and (ii) protein complexes that are non-DNA sequence-specific and modify nucleosome or remodel chromatin (Sinha et al. Citation2005). Previous studies have shown that, under the stimulation of serum, transport of BRD2 protein into the fibroblast nucleus participates in the nuclear protein complex, including E2F protein, and transactivates the promoters of E2F-dependent mammalian cell cycle genes, including dihydrofolate reductase, cyclins A, D1 and E (Guo et al. Citation2000). Further studies have confirmed that BRD2 protein binds to the promoter of cyclin A, and its overexpression promotes the transcriptional activity of histone H4 and related cyclin-dependent kinase activity, to accelerate the cell cycle (Sinha et al. Citation2005). As a cell cycle-induced transcriptional activator, BRD2 promotes the activation of NF-κB and the expression of downstream inflammatory genes in microglia when subjected to lipopolysaccharide challenge (Demars et al. Citation2018).
Among the cellular proteins screened with chBRD2 protein as a ‘bait’ protein, Caprin-1 is a kind of RNA-binding protein that can affect cell survival and growth by selectively binding to a variety of genes involved in cell growth, differentiation and migration, including c-Myc and cyclin D2 (Yang et al. Citation2019). Drebrin (DBN1) is a major F-actin binding protein in dendritic spines that are critically involved in the regulation of dendritic spine morphogenesis, pathology, and plasticity (Yamazaki et al. Citation2018). Charged multivesicular body protein 1 (CHMP1) is a nuclear matrix protein affecting chromatin structure and cell-cycle progression (Stauffer et al. Citation2001). Transcription factor 12 (TCF12) is responsible for cell development and differentiation and plays a key role in the differentiation of lymphocytes and the development of neural and mesenchymal tissues (Gao et al. Citation2019). Furthermore, the protein complex formed by RUNX1 and BRD2 participates in the cellular decision about the restricted (R)-point of division or death (Lee et al. Citation2019).
chBRD2 and virus proliferation
Recognition of acetylated chromatin by BET proteins is a hallmark for transcriptional activation and anchoring viral genomes to mitotic chromosomes of the host. In recent years, BRD2 protein has been shown to regulate the expression level of viral and host genes through interactions with viral proteins. For example, during the incubation period of Kaposi’s sarcoma-associated herpes virus (KSHV), BRD2 interacts with the C-terminal chromatin binding domain of latency-associated nuclear antigen 1 (LANA-1), which binds the virus genome to the cell chromosome, thus promoting the persistence of the KSHV virus and participating in the transcriptional regulation and maintenance of viral and cellular genes in vitro (Viejoborbolla et al. Citation2003, Citation2005). The ET domain of the BRD2 protein can be integrated into the host genome by the interaction between the ET domain of BRD2 protein and the γ-porcine endogenous retrovirus A/C integrase (PERV IN) (Gallay et al. Citation2019). Recent studies have shown that both BET inhibitors and RNA interference with BRD4, a member of the BRD2 family, can inhibit the proliferation of the virus, and such inhibition does not affect the transcription of the above-mentioned viral genes, but rathe inhibits the adsorption stage between the virus and the host cell to inhibit replication (Wang et al. Citation2020). At the same time, inhibition of BRD4 can activate antiviral innate immunity which is dependent on interferon regulatory factor 3 (IRF3) (Wang et al. Citation2020).
The current study identified some cellular proteins that may interact with chBRD2 protein and participate in virus invasion and replication by Co-IP combined with LC-MS. For instance, HIV-1 uses host factor RUVBL2 to balance the expression of viral proteins (Mu et al. Citation2015). RUVBL2 is involved in viral gene transcription of hepatitis B virus (Nishitsuji et al. Citation2018), influenza A virus (Kakugawa et al. Citation2009) and Ebola virus (Morwitzer et al. Citation2019). Machupo virus, which causes haemorrhagic fever, enters human cells through the binding of its envelope glycoprotein to transferrin receptor 1 (TFRC) (Sjöström et al. Citation2021). SRSF protein kinases 1 and 2 are essential to host factors for human coronaviruses including SARS-CoV-2 (Heaton et al. Citation2020).
Interaction between chBRD2 and chMATR3 in the nucleus
Previous research found that chBRD2 or chMATR3 proteins may interact with NDV matrix protein using yeast two-hybrid system (Duan et al. Citation2018). Recently, it was reported that the ET domain of the chBRD2 protein interacted with NDV matrix protein, and small interference RNA-mediated chBRD2 gene knockout or chBRD2 gene overexpression significantly enhanced or decreased NDV replication by up- or down-regulating viral RNA synthesis and transcription, respectively (Duan et al. Citation2020). As one of the first 12 reported nuclear matrix proteins, MATR3 plays an important role in the nuclear matrix, which participates in the nuclear matrix and provides the structural framework for nuclear activities and is closely related to the functions of nuclear and gene transcription regulation, mRNA precursor splicing, stability, DNA damage repair and cell proliferation (Coelho et al. Citation2015). It has been shown that MATR3 interacts with bromodomain adjacent to zinc finger domain protein 1A (BAZ1A) to participate in chromatin remodelling (Zeitz et al. Citation2009). A previous study found that NDV matrix protein interacted with chMATR3 in the nucleus, and decreased expression of chMATR3 gene significantly increased replication efficiency and pathogenicity of NDV (Deng Citation2019). In addition, the transcription of the chMATR3 or chBRD2 gene was obviously decreased in both NDV-infected cells and pEGFP-M-transfected cells in a dose-dependent manner, which was of benefit for both NDV RNA synthesis and transcription (Deng Citation2019; Duan et al. Citation2020). However, further studies are needed to explore potential roles and interactions in the life cycle of NDV.
Conclusions
In total, 225 identified proteins were found in chBRD2 complexes by Co-IP combined with LC-MS analysis. GO analysis showed that these proteins played key roles in gene transcriptional regulation, cell cycle and development, immunity and viral infection. KEGG pathway analysis revealed that these proteins were mainly involved in genetic information processing, immunity, cellular processes and translation. One targeted cellular protein chMATR3 was confirmed to interact with chBRD2 using fluorescence co-localisation and Co-IP assays. These findings provide new information regarding the functions of chBRD2 and targets for further research of NDV replication and pathogenesis.
Table S3. The cellular proteins interacting with chBRD2 using Vene analysis.
Download MS Excel (29.8 KB)Table S2. The cellular proteins interacting with GFP-chBRD2.
Download MS Excel (45.1 KB)Table S1. The cellular proteins interacting with GFP.
Download MS Excel (28.8 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Belkina, A. C., and G. V. Denis. 2012. “BET Domain Co-regulators in Obesity, Inflammation and Cancer.” Nature Reviews Cancer 12: 465–477. doi:https://doi.org/10.1038/nrc3256.
- Chiang, C. M. 2009. “Brd4 Engagement from Chromatin Targeting to Transcriptional Regulation: Selective Contact with Acetylated Histone H3 and H4.” F1000 Medicine Reports 1: 98. doi:https://doi.org/10.3410/B1-98.
- Coelho, M., J. Attig, N. Bellora, J. König, M. Hallegger, M. Kayikci, E. Eyras, J. Ule, and C. Smith. 2015. “Nuclear Matrix Protein Matrin3 Regulates Alternative Splicing and Forms Overlapping Regulatory Networks with PTB.” The EMBO Journal 34: 653–668. doi:https://doi.org/10.15252/embj.201489852.
- Demars, K. M., C. Yang, C. I. Castro-Rivera, and E. Candelario-Jalil. 2018. “Selective Degradation of BET Proteins with dBET1, a Proteolysis-targeting Chimera, Potently Reduces Pro-inflammatory Responses in Lipopolysaccharide-activated Microglia.” Biochemical and Biophysical Research Communications 497: 410–415. doi:https://doi.org/10.1016/j.bbrc.2018.02.096.
- Deng, S. S. 2019 “Study on the Role of Chicken Matrix 3 Protein in the Replication of Newcastle Disease Virus” Mater Dissertation Research Paper. Guizhou University. Guiyang. China. https://kns.cnki.net/kcms/detail/detail.aspx?dbcode=CMFD&dbname=CMFD201902&filename=1019846848.nh&v=liPTyOHyxxKG90zTaw18CZ0OvCwsPysCM79vSCy324i%25mmd2BcND%25mmd2BBoUUAbdXyYwFI3ux.
- Denis, G. V., M. E. Mccomb, D. V. Faller, A. Sinha, P. B. Romesser, and C. E. Costello. 2006. “Identification of Transcription Complexes that Contain the Double Bromodomain Protein Brd2 and Chromatin Remodelling Machines.” Journal of Proteome Research 5: 502–511. doi:https://doi.org/10.1021/pr050430u.
- Duan, Z. Q., Y. F. Han, L. Zhou, C. Yuan, Y. B. Wang, C. Q. Zhao, H. Tang, and J. Q. Chen. 2020. “Chicken Bromodomain-containing Protein 2 Interacts with the Newcastle Disease Virus Matrix Protein and Promotes Viral Replication.” Veterinary Research 51: 120. doi:https://doi.org/10.1186/s13567-020-00846-1.
- Duan, Z. Q., H. X. Xu, X. Q. Ji, J. F. Zhao, H. Q. Xu, Y. Hu, S. S. Deng, S. L. Hu, and X. F. Liu. 2018. “Importin α5 Negatively Regulates Importin β1-mediated Nuclear Import of Newcastle Disease Virus Matrix Protein and Viral Replication and Pathogenicity in Chicken Fibroblasts.” Virulence 9: 783–803. doi:https://doi.org/10.1080/21505594.2018.1449507.
- Florence, B., and D. Faller. 2001. “You Bet-cha: A Novel Family of Transcriptional Regulators.” Frontiers in Bioscience: A Journal and Virtual Library 6: D1008- D1018. doi:https://doi.org/10.2741/florence.
- Gallay, K., G. Blot, M. Chahpazoff, H. Yajjouhamalian, M. Confort, C. D. Boisseson, A. Leroux, et al. 2019. “In Vitro, in Cellulo and Structural Characterizations of the Interaction between the Integrase of Porcine Endogenous Retrovirus A/C and Proteins of the BET Family.” Virology 532: 69–81. doi:https://doi.org/10.1016/j.virol.2019.04.002.
- Gao, S., T. Bian, Y. Zhang, M. Su, and Y. Liu. 2019. “TCF12 Overexpression as a Poor Prognostic Factor in Ovarian Cancer.” Pathology, Research and Practice 215: 152527. doi:https://doi.org/10.1016/j.prp.2019.152527.
- Gittings, L. M., S. C. Foti, B. C. Benson, P. Gami-patel, A. M. Isaacs, and T. Lashley. 2019. “Heterogeneous Nuclear Ribonucleoproteins R and Q Accumulate in Pathological Inclusions in FTLD-FUS.” Acta Neuropathologica Communications 7: 18. doi:https://doi.org/10.1186/s40478-019-0673-y.
- Guo, N., D. V. Faller, and G. V. Denis. 2000. “Activation-induced Nuclear Translocation of RING3.” Journal of Cell Science 113: 3085–3091. doi:https://doi.org/10.1080/152165400300001499.
- Gursoyyuzugullu, O., C. Carman, and B. D. Price. 2017. “Spatially Restricted Loading of BRD2 at DNA Double-strand Breaks Protects H4 Acetylation Domains and Promotes DNA Repair.” Scientific Reports 7: 12921. doi:https://doi.org/10.1038/s41598-017-13036-5.
- Han, S. P., Y. H. Tang, and R. Smith. 2010. “Functional Diversity of the hnRNPs: Past, Present and Perspectives.” The Biochemical Journal 430: 379–392. doi:https://doi.org/10.1042/bj20100396.
- Heaton, B. E., J. D. Trimarco, C. E. Hamele, A. T. Harding, A. Tata, X. Zhu, P. R. Tata, C. M. Smith, and N. S. Heaton. 2020. “SRSF Protein Kinases 1 and 2 are Essential Host Factors for Human Coronaviruses Including SARS-CoV-2.” bioRxiv. doi:https://doi.org/10.1101/2020.08.14.251207.
- HnilicovÁ, J., S. Hozeifi, E. StejskalovÁ, E. DuškovÁ, I. Poser, J. HumpolÍčkovÁ, M. Hof, and D. Staněk. 2013. “The C-terminal Domain of Brd2 Is Important for Chromatin Interaction and Regulation of Transcription and Alternative Splicing.” Molecular Biology of Cell 24: 3557–3568. doi:https://doi.org/10.1091/mbc.E13-06-0303.
- Idauden, M., A. López-perrote, and O. Llorca. 2020. “RUVBL1-RUVBL2 AAA-ATPase: A Versatile Scaffold for Multiple Complexes and Functions.” Current Opinion in Structural Biology 67: 78–85. doi:https://doi.org/10.1016/j.sbi.2020.08.010.
- Kakugawa, S., M. Shimojima, G. Neumann, H. Goto, and Y. Kawaoka. 2009. “RuvB-like Protein 2 Is A Suppressor of Influenza A Virus Polymerases.” Journal of Virology 83: 6429–6434. doi:https://doi.org/10.1128/jvi.00293-09.
- Kanno, T., Y. Kanno, R. M. Siegel, M. Jang, M. J. Lenardo, and K. Ozato. 2004. “Selective Recognition of Acetylated Histones by Bromodomain Proteins Visualized in Living Cells.” Molecular Cell 13: 33–43. doi:https://doi.org/10.1016/s1097-2765(03)00482-9.
- Lee, J., T. Park, and S. Bae. 2019. “Involvement of RUNX and BRD Family Members in Restriction Point.” Molecules and Cells 42: 836–839. doi:https://doi.org/10.14348/molcells.2019.0256.
- Leroy, G., R. Rickards, and S. J. Flint. 2008. “The Double Bromodomain Proteins Brd2 and Brd3 Couple Histone Acetylation to Transcription.” Molecular Cell 30: 51–60. doi:https://doi.org/10.1016/j.molcel.2008.01.018.
- Lu, J., N. Bergman, N. Sadri, and R. J. Schneider. 2006. “Assembly of AUF1 with eIF4G-poly (A) Binding Protein Complex Suggests a Translation Function in AU-rich mRNA Decay.” RNA (New York, N.Y.) 12: 883–893. doi:https://doi.org/10.1261/rna.2308106.
- Morwitzer, M., S. Tritsch, L. Cazares, M. Ward, J. Nuss, S. Bavari, and S. Reid. 2019. “Identification of RUVBL1 and RUVBL2 as Novel Cellular Interactors of the Ebola Virus Nucleoprotein.” Viruses 11: 372. doi:https://doi.org/10.3390/v11040372.
- Mu, X., Y. Fu, Y. Zhu, X. Wang, Y. Xuan, H. Shang, S. Goff, and G. Gao. 2015. “HIV-1 Exploits the Host Factor RuvB-like 2 to Balance Viral Protein Expression.” Cell Host & Microbe 18: 233–242. doi:https://doi.org/10.1016/j.chom.2015.06.018.
- Nishitsuji, H., S. Ujino, K. Harada, and K. Shimotohno. 2018. “TIP60 Complex Inhibits Hepatitis B Virus Transcription.” Journal of Virology 92: e01788–17. doi:https://doi.org/10.1128/jvi.01788-17.
- Peng, J. H., W. Dong, L. Chen, T. T. Zou, Y. P. Qi, and Y. L. Liu. 2007. “Brd2 Is a TBP-associated Protein and Recruits TBP into E2F-1 Transcriptional Complex in Response to Serum Stimulation.” Molecular and Cellular Biochemistry 294: 45–54. doi:https://doi.org/10.1007/s11010-006-9223-6.
- Pham, T. X., M. Bae, Y. Lee, Y. Park, and J. Lee. 2018. “Transcriptional and Posttranscriptional Repression of Histone Deacetylases by Docosahexaenoic Acid in Macrophages.” The Journal of Nutritional Biochemistry 57: 162–169. doi:https://doi.org/10.1016/j.jnutbio.2018.03.002.
- Rahman, S., M. E. Sowa, M. Ottinger, J. A. Smith, Y. Shi, J. W. Harper, and P. M. Howley. 2011. “The Brd4 Extraterminal Domain Confers Transcription Activation Independent of pTEFb by Recruiting Multiple Proteins, Including NSD3.” Molecular and Cellular Biology 31: 2641–2652. doi:https://doi.org/10.1128/mcb.01341-10.
- Sansam, C. G., K. Pietrzak, B. Majchrzycka, M. A. Kerlin, J. Chen, S. Rankin, and C. L. Sansam. 2018. “A Mechanism for Epigenetic Control of DNA Replication.” Genes Development 32: 224–229. doi:https://doi.org/10.1101/gad.306464.117.
- Sihn, C. R., S. Y. Cho, J. H. Lee, T. R. Lee, and S. H. Kim. 2007. “Mouse Homologue of Yeast Prp19 Interacts with Mouse SUG1, the Regulatory Subunit of 26S Proteasome.” Biochemical and Biophysical Research Communications 356: 175–180. doi:https://doi.org/10.1016/j.bbrc.2007.02.134.
- Sinha, A., D. V. Faller, and G. V. Denis. 2005. “Bromodomain Analysis of Brd2-dependent Transcriptional Activation of Cyclin A.” The Biochemical Journal 387: 257–269. doi:https://doi.org/10.1042/bj20041793.
- SjÖstrÖm, D. J., A. Lundgren, S. J. Garforth, and S. Bjelic. 2021. “Tuning the Binding Interface between Machupo Virus Glycoprotein and Human Transferrin Receptor.” Proteins 89: 311–321. doi:https://doi.org/10.1002/prot.26016.
- Stauffer, D. R., T. L. Howard, T. Nyun, and S. M. Hollenberg. 2001. “CHMP1 Is a Novel Nuclear Matrix Protein Affecting Chromatin Structure and Cell-cycle Progression.” Journal of Cell Science 114: 2383–2393. doi:https://doi.org/10.1242/jcs.114.13.2383.
- Taniguchi, Y. 2016. “The Bromodomain and Extra-Terminal Domain (BET) Family: Functional Anatomy of BET Paralogous Proteins.” International Journal of Molecular Sciences 17: 1849–1873. doi:https://doi.org/10.3390/ijms17111849.
- Tomohiko, K., K. Yuka, L. Gary, E. I. Campos, H. W. Sun, S. R. Brooks, G. Vahedi, T. D. .Heightman, B. A. Garcia, and R. Danny. 2014. “BRD4 Assists Elongation of Both Coding and Enhancer RNAs by Interacting with Acetylated Histones.” Nature Structural & Molecular Biology 21: 1047–1057. doi:https://doi.org/10.1038/nsmb.2912.
- Umehara, T., Y. Nakamura, M. K. Jang, K. Nakano, A. Tanaka, K. Ozato, B. Padmanabhan, and S. Yokoyama. 2010. “Structural Basis for Acetylated Histone H4 Recognition by the Human BRD2 Bromodomain.” Journal of Biological Chemistry 285: 7610–7618. doi:https://doi.org/10.1074/jbc.M109.062422.
- Viejoborbolla, A., E. Kati, J. A. Sheldon, K. Nathan, K. Mattsson, L. Szekely, and T. F. Schulz. 2003. “A Domain in the C-terminal Region of Latency-associated Nuclear Antigen 1 of Kaposi’s Sarcoma-associated Herpesvirus Affects Transcriptional Activation and Binding to Nuclear Heterochromatin.” Journal of Virology 77: 7093–7100. doi:https://doi.org/10.1128/jvi.77.12.7093-7100.2003.
- Viejoborbolla, A., M. Ottinger, E. Bruning, A. Burger, R. Konig, E. Kati, J. Sheldon, and T. F. Schulz. 2005. “Brd2/RING3 Interacts with a Chromatin-binding Domain in the Kaposi’s Sarcoma-associated Herpesvirus Latency-associated Nuclear Antigen 1 (LANA-1) that Is Required for Multiple Functions of LANA-1.” Journal of Virology 79: 13618–13629. doi:https://doi.org/10.1128/jvi.79.21.13618-13629.2005.
- Wang, J., G.-L. Li, S.-L. Ming, C.-F. Wang, L.-J. Shi, B.-Q. Su, H.-T. Wu, et al. 2020. “BRD4 Inhibition Exerts Anti-viral Activity through DNA Damage-dependent Innate Immune Responses.” PLOS Pathogens 16 (3): e1008429. doi:https://doi.org/10.1371/journal.ppat.1008429.
- Yamazaki, H., Y. Sasagawa, H. Yamamoto, H. Bito, and T. Shirao. 2018. “CaMKIIβ Is Localized in Dendritic Spines as Both Drebrin-dependent and Drebrin-independent Pools.” Journal of Neurochemistry 146: 145–159. doi:https://doi.org/10.1111/jnc.14449.
- Yang, Z., H. Qing, H. Gui, J. Luo, L. Dai, and B. Wang. 2019. “Role of Caprin-1 in Carcinogenesis.” Oncology Letters 18: 15–21. doi:https://doi.org/10.3892/ol.2019.10295.
- Zang, K., J. Wang, M. Dong, R. Sun, Y. Wang, Y. Huang, X. Liu, Y. Li, F. Wang, and M. Yu. 2013. “Brd2 Inhibits Adipogenesis via the ERK1/2 Signaling Pathway in 3T3-L1 Adipocytes.” PloS One 8: e78536. doi:https://doi.org/10.1371/journal.pone.0078536.
- Zeitz, M., K. Malyavantham, B. Seifert, and R. Berezney. 2009. “Matrin 3: Chromosomal Distribution and Protein Interactions.” Journal of Cellular Biochemistry 108: 125–133. doi:https://doi.org/10.1002/jcb.22234.
- Zeng, L., and M. M. Zhou. 2002. “Bromodomain: An Acetyl-lysine Binding Domain.” FEBS Letters 513: 124–128. doi:https://doi.org/10.1016/s0014-5793(01)03309-9.