Abstract
Amplified fragment length polymorphism (AFLP) marker and cytological analyses were employed to evaluate the genetic relationships and diversity of Chinese snakegourd. According to the cytological analyses, accessions 1, 2 and 3 were diploid (2n = 22), and accessions 4 to 12 were octaploid (2n = 88). Fourteen AFLP primer combinations generated a total of 645 reproducible loci among 12 Chinese snakegourd accessions studied, of which 628 loci were polymorphic with an average of 44.9 polymorphic bands per primer combination. Genetic similarities were obtained using Nei and Li similarity coefficients, and a dendrogram reflecting the relationship of 12 accessions was made using the unweighted pair-group method with arithmetic averages (UPGMA) clustering method. Based on the cluster analyses, 12 accessions were classified into two primary groups, in which group one included accessions 1, 2 and 3 that were diploid, and the other nine octaploid accessions formed group two. Analysis of molecular variance (AMOVA) revealed much greater variation among groups than within them. Meanwhile, some genotypes that might have great medicinal potential were also proposed according to their genetic relationships. These results provide exact evaluation of the relationships among Trichosanthes kirilowii accessions and give useful information for further investigation and utilization of this species.
Introduction
Chinese snakegourd (Trichosanthes kirilowii Maxim), belonging to the genus Trichosanthes in the Cucurbitaceae family, is a woody perennial plant which is distributed worldwide due to its great adaptation to different environments. It is widely cultivated in Southeast Asia and North Australia. In China, T. kirilowii is reputedly used in traditional Chinese medicine for the treatment of inflammation, cough and phlegm, and has more than two thousand years of cultivation history (Tang & Eisenbrand Citation1992). The medical value of seed, fruit peel, and root of T. kirilowii has been studied and applied in antitumor, anti-HIV, and antityrosinase therapy (Yang et al. Citation2007; Shin et al. Citation2008; Takahashi et al. Citation2009). Most T. kirilowii were wild types that were cross-pollinated under no control. The identification of cultivars is difficult due to the plant’s complicated and changing morphologies, so pharmacists classify this herbal medicine empirically based on its producing area. Although T. kirilowii has been commercially cultivated in some of its distribution centers, a large proportion of T. kirilowii was harvested from wild land. Some species with differing medicinal usage are difficult to identify because of similar morphology. One taxonomist described it as “this species had the most intractable taxonomic problem in Eastern Asia Cucurbitaceae center” (Jeffrey, Citation1980). Until now, little systematic research of genetic diversity and phylogenetic relationships has been carried out with Chinese snakegourd.
Molecular markers, used for morphological and physiological characterization of plant species, have provided a powerful approach to analyze genetic relationships among species at the DNA level (Manifesto et al. Citation2001). Amplified fragment length polymorphism (AFLP) is a novel polymerase chain reaction (PCR)-based assay for plant DNA fingerprinting which reveals significant levels of DNA polymorphism (Vos et al. Citation1995). AFLP markers are randomly distributed throughout the genome and were reported to be efficient and reliable in supporting the low level taxonomy (Kumar Citation1999; Guthridge et al. Citation2001).
In the present study we identified the chromosome ploidy levels of 12 T. kirilowii accessions, which were selected from different production areas. The AFLP technique was applied to analyze the genetic diversities and phylogenetic relationships among 12 T. kirilowii accessions. This work will facilitate the exact identification of certain accessions of the T. kirilowii germplasms, and widen the genetic basis of breeding material selection.
Materials and methods
Twelve typical T. kirilowii accessions were collected from six traditional productive areas in China. The relevant information about these accessions is listed in Table . All materials were grown in subdivided areas at the farm of Zhejiang University. The morphologies of leaf and mature fruit were recorded in the same growing stage (June 2006).
Table 1. T. kirilowii accessions used in this study.
Vigorously growing root tips were collected from regenerated plants of 12 accessions. To accumulate metaphase cells, the root tips were pretreated with 2 mM 8-hydroxyquinoline for 3–4 h. The root tips were then fixed in 3:1 ethanol/acetic acid. Chromosome observation procedure was based on the method of Wu et al. (Citation1997).
Plant DNA was extracted using the cetyltrimethyl ammonium bromide (CTAB) method according to Doyle & Doyle (Citation1990). AFLPs were developed using a scaled-down modification of the originally described procedure (Vos et al. Citation1995). Fourteen primer combinations with high polymorphic expression were selected from 64 primer combinations, and the sequence of them are listed in Table .
Table 2. The number of polymorphism bands detected by 14 primer combinations. Primer sequences are also listed.
Genomic DNA (250 ng) was digested with three units each of Mse I and EcoR I endonuclease at 37°C for 3 h, and then 70°C for 15 min. The digested DNA fragments were ligated to Mse I and EcoR I adaptors with T4 DNA ligase for 2 h at 20°C. The ligated DNA was diluted to 1:10 in TE buffer (10 mM Tris-HCL (PH 8.0), 0.1 mM EDTA) and stored at –20°C. PCR was performed in two consecutive reactions: pre-selective and selective PCR. In the pre-selective reaction, genomic DNA was amplified using an AFLP primer pair, each having one selective nucleotide. Accordingly, 5 μl of diluted ligation product, 2 μl each of Mse I (25 ng μl–1) and EcoR I (25 ng μl–1), 2 μl dNTP (2 mM), 2 μl 10 × PCR buffer mixed with MgCl2 (15 mM) for AFLP and 0.4 unit of Taq polymerase were mixed for the pre-selective reaction in a total volume of 20 μl. The pre-selective reactions were performed as follows: 24 cycles of 30 s at 94°C, 60 s at 56°C, 60 s at 72°C. Pre-selective PCR amplification was confirmed by gel electrophoresis and the amplified product was diluted to 1:50 in TE buffer and used as a template for the selective amplification using AFLP primers, each containing three selective nucleotides. Selective PCR amplification was performed in 20 μl reactions consisting of 5 μl pre-selective template DNA, 2 μl EcoR I (10 ng μl–1), 6 μl Mse I (10 ng μl–1), 2 μl dNTP (2 mM), 2 μl 10 × PCR buffer mixed with MgCl2 (15 mM) for AFLP and 1 unit of Taq polymerase. Selective PCR amplification reactions were performed for one cycle at 94°C for 30 s, 65°C for 60 s, 72°C for 60 s. Then the annealing temperature was lowered 0.7°C during the first 12 cycles down to 56°C. The amplification products (15 μl) were mixed with 8 μl loading buffer, denatured at 94°C for 10 min, and quickly cooled in ice slurry. Then 4 μl were taken and electrophoresed at constant power (75 W) in a 0.4 mm thick, 6% denaturing polyacrylamide gel with 1 × TBE running buffer until the blue band reached 3/4 of the gel, as described by Sambrook et al. (Citation1989). Analysis was carried out by silver staining (Promega Corporation, Madison, USA) and overnight drying before being photographed.
Details of molecular weight of PCR products were collected and identified. Bands clearly visible in at least one cultivar were scored (1 for present, 0 for absent) and entered into a data matrix to create a qualitative data matrix. The fragment’s size was estimated by interpolation from the migration distance of marker fragments.
Similarities based on the relationship between the genotypes were presented in the form of a dendrogram, according to the resulting similarity matrix, developed by following unweighted pair group method with arithmetic mean algorithm (UPGMA), using SAHN clustering analysis of NTSYS-PC, version 2.1 (Rohlf Citation1994). Pair-wise similarities between accessions were calculated using Jaccard’s coefficient for qualitative data (Jaccard Citation1908), according to the formula: Jaccard’s coefficient = a/(n – d), where n is the total number of polymorphic bands, a is the bands present in both accessions and d is the bands absent in both accessions. Estimates of genetic similarity among all genotypes were also calculated using the Nei & Li (Citation1979) coefficient of similarity between two individuals (i and j), according to the formula: Nei & Li’s coefficient = 2a/(b + c), where a is the number of shared bands present in both samples i and j; b is the total number of bands of individual i, and c is the total number of bands of individual j. At first, the goodness-of-fit of the clustering was tested using the MXCOMP program, which directly compares the original similarity matrix and the cophenetic value matrix, as suggested by Rohlf (Citation1994). Then dendrograms were constructed by NTSYS-PC program.
To estimate the partitioning of AFLP genotypic variation between and within groups, analysis of molecular variance (AMOVA) (Excoffier et al. Citation1992) was also performed, using Arlequin software version 2000 (Schneider et al. Citation2000).
Results
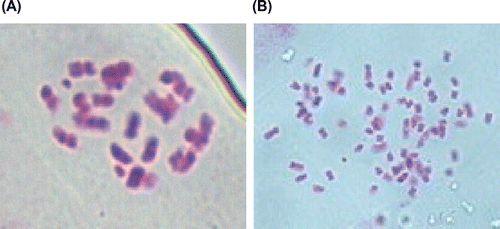
The ploidy level of each accession was measured using chromosomes of the dividing cell in metaphase. Accessions 1, 2 and 3 were determined to be diploid (2n = 22), accessions 4 to 12 were octaploid (2n = 88) (Figure ). The leaf and mature fruit configuration of 12 T. kirilowii accessions were recorded at the same growth stage and the leaf shape, and fruit configuration, and color showed large differences (supplementary Figures s and s.).
Figure s1 (Colour online) Leaf morphologies of the 12 T. kirilowii accessions. The numbers on the pictures are in accordance with the numbers in the first line of Table .
Figure s2 (Colour online) Mature fruit morphologies of the 12 T. kirilowii accessions. The numbers on the pictures are in accordance with the numbers in the first line of Table .

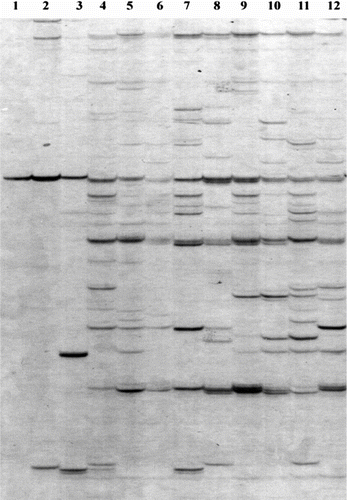
The detailed data for the AFLP analysis of 12 accessions of T. kirilowii are summarized in Table . The 14 pairs of primer generated a total of 645 scorable fragments of which 628 bands were polymorphic with an average of 44.9 polymorphic bands per primer combination. The primer combinations differed in their ability to detect polymorphism within populations. The total number of polymorphic bands over all accessions detected by individual primer pair varied from 29 (primer combination E-AGG/M-CTG) to 69 (primer combination E-ATG/M-CAG). The AFLP result of primer combination E-ACG/M-CAA has the high-test Nei’s (Citation1973) gene diversity and Shannon’s Information index: 0.474 and 0.667, respectively (data are not shown). This indicated that this primer combination had the highest ability to distinguish the whole genotypes of T. kirilowii. Part of the AFLP bands amplified by primer combination E-ATG/M-CAG is presented in Figure .
Figure 2 (Colour online) AFLP profiles of the 12 T. kirilowii accessions generated by primer combination E-ATG/M-CAG. The numbers in the lanes correspond to the accession numbers of Table .
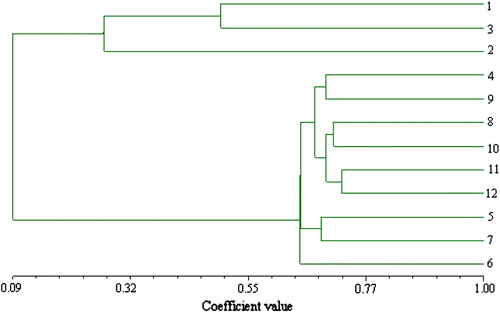
The AFLP data generated from 628 fragments over 14 primer pairs were used to calculate the similarity coefficients for all the 12 accessions, and dendrograms were developed using the UPGMA method by NTSYS-PC version 2.1. Similarity matrixes based on Jaccard, and Nei & Li coefficients were generated, respectively. The dendrogram had a matrix correlation r = 0.9, which is interpreted as a good fit. So, only the Jaccard matching data were used for generating the dendrogram presented here (Figure ). Twelve accessions of T. kirilowii were clustered into two main groups at a coefficient value of 0.26 (Figure ). One group is comprised of three diploid accessions, 1, 2 and 3, and the other nine octaploid types of T. kirilowi formed group two. In group one, accessions 1 and 3 were grouped into one minor cluster at a coefficient value of 0.51. The nine octaploid accessions were clustered into three minor groups. Accession 6 was found to be the most divergent line and separated from the other accessions at a coefficient value of 0.65 in main group two. At the 0.69 level, accessions 5 and 7 formed one minor group. Another minor group was composed of three rami. Accessions 11 and 12 were clustered into one ramus at a coefficient value of 0.73; accessions 8 and 10 formed another ramus at the coefficient value of 0.74; accessions 4 and 9 composed the third ramus.
Figure 3 The dendrogram of the 12 T. kirilowii accessions generated by UPGMA from the similarity matrix of AFLP data (calculated with Jaccard’s method). The number in the figure corresponds to the accession number of Table .
AMOVA results showed that most of the variations (99.67%) were found among groups, and variations within the groups were quite small (0.33%). Both variations were highly significant (p < 0.001) (Table ).
Table 3. AMOVA analyses among and within groups.
Discussion
Nowadays, T. kirilowii has been given extensive attention due to its medicinal value in China. However, the classification of this species was considered the most intractable taxonomic problem in the center of Cucurbitaceae diversity in Eastern Asia (Jeffrey Citation1980). The object of our study was to investigate the phylogenetic relationship and cytogenetic traits of T. kirilowii from different areas of production in China.
Cytological analysis indicated that of T. kirilowii accessions have a base chromosome number of x = 11, and all of them were diploid (2n = 22), tetraploid (2n = 44) or octoploid (2n = 88) (Darlington & Wylie Citation1955). As we know, chromosome variation is directly related to the evolution of the plant. In our results, accessions 1, 2 and 3 were diploid, and all the other nine accessions were octoploid. According to the cluster analysis of AFLP markers, diploid accessions were clustered into one group and octoploid accessions were separated into another group. All nine octoploid T. kirilowii studied were used as herbal medicine. Genetic variance between the diploid and octoploid groups measured by AMOVA was 99.67% of the total variance. This high level of variance implied that octoploids suffered great genetic alteration in their long evolutionary history, and formed the stable polyploidy of T. kirilowii. Polyploidy comes from whole-genome duplication and comes at the cost of major genomic instability (Mayer & Aguilera Citation1990), which persists until the genome returns to functionally normal ploidy through mutation, gene loss or genomic rearrangements. However, genomic duplication has been proposed as an advantageous path to evolutionary innovation (Ohno Citation1970), because duplicated genes can supply genetic raw material for the emergence of new functions through the forces of mutation and natural selection. In principle, coordinated duplication of an entire genome may allow for large-scale adaptation to new environments. This may be the reason why most of the T. kirilowii were octoploid.
Based on the phylogenetic analysis from AFLP data, the minimum genetic similarity was observed between the accessions of 11 and 12, which were both collected from Jiaozuo, Henan Province. In addition, edible seed accessions 2 and 3 were clustered into one group, and both were collected from Changxing, Zhejiang Province. However, the relationship among other T. kirilowii accessions was not obviously related to their geographical distribution. For example, accession 4 from Pinghu, Zhejiang Province (south China), and accession 9 from Anyang, Henan Province (middle north of China) were clustered together. Accession 5 from Zhangqiu, Shandong province (north China), and accession 7 from Puyang, Henan Province (middle north of China) showed a close relationship as well. This may be because the present accession of T. kirilowii was grown in different locations under no environmental control, and no cultivar selection pressure could induce DNA sequence change.
Accessions 1, 4, 6, 7, 8, 11, and 12 were collected from a herbal medicine production farm and had been used as herbal medicines for a long period of time in the local area. The function of the three wild type accessions 5, 9, and 10 were unknown, however, they showed close relationships with other medicinal accessions. These three accessions might have potential medicinal value. For further utilization and investigation of this traditional herbal plant, chemical component analysis of each accession needs to be performed.
Acknowledgments
The authors are grateful to the Agricultural Bureau of Changxing Country for providing us with three T. kirilowii accessions. We are also grateful to master Fan Tai-Wei for valuable advice on data analysis.
References
- Darlington , CD and Wylie , AP . 1955 . Chromosone atlas of flowering plants , London : Allen and Unwin .
- Doyle , JJ and Doyle , JL . 1990 . Isolation of plant DNA from fresh tissue . Focus , 12 : 13 – 15 .
- Excoffier , L , Smouse , PE and Quattro , JM . 1992 . Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data . Genetics , 131 : 479 – 491 .
- Guthridge , KM , Dupal , MP , Kolliker , R , Jones , ES , Smith , KF and Forster , JW . 2001 . AFLP analysis of genetic diversity within and between populations of perennial ryegrass (Lolium perenne L.) . Euphytica , 122 : 191 – 201 .
- Jaccard , P . 1908 . Nouvelles recherches sur la distribution florale . Bull Soc Vaudense Sci Nat , 44 : 223 – 270 .
- Jeffrey C. 1980. The Cucurbitaceae of Eastern Asia. Kew, Richmond, UK: Royal Botanic Gardens. p. 38–52.
- Kumar , LS . 1999 . DNA markers in plant improvement: An overview . Biotechnol Adv , 17 : 143 – 182 .
- Manifesto , MM , Schlatter , AS , Hopp , HE , Suarez , EY and Dubcovky , J . 2001 . Quantitative evaluation of genetic diversity germplasm using molecular markers . Crop Sci , 41 : 682 – 690 .
- Mayer , VW and Aguilera , A . 1990 . High levels of chromosome instability in polyploids of saccharomyees cerevisiae . Mutat Res , 231 : 177 – 186 .
- Nei , M . 1973 . Analysis of gene diversity in subdivided populations . P Natl Acad Sci USA , 70 : 3321 – 3323 .
- Nei , M and Li , WH . 1979 . Mathematical model for studying genetic variation in terms of restriction endonucleases . P Natl Acad Sci USA , 76 : 5269 – 5273 .
- Ohno , S . 1970 . Evolution by gene duplication , London : George Allen and Unwin . p. 160
- Rohlf FJ. 1994. NTSYS-pc, Numerical Taxonomy and Multivariate Analysis System Version 2.0. Exeter Software. New York: Setauket.
- Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning. a laboratory manual. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press. p. 11–47.
- Schneider S, Roessli D, Excoffier L. 2000. Arlequin: A Software for Population Genetics Data Analysis, Version 2.0. Genetics and biometry Laboratory, Department of Anthropology, University of Geneva, Switzerland.
- Shin , JW , Son , JY , Kang , JK , Han , SH , Cho , CK and Son , CGJ . 2008 . Trichosanthes kirilowii tuber extract induces G2/M phase arrest via inhibition of tubulin polymerization in HepG2 cells . J Ethnopharmacol , 115 : 209 – 216 .
- Takahashi , N , Yoshida , Y , Sugiura , T , Matsuno , K , Fujino , A and Yamashita , U . 2009 . Cucurbitacin D isolated from Trichosanthes kirilowii induced apoptosis in human hepatocellular carcinoma cells in vitro . Int Immunopharmacol , 9 : 508 – 513 .
- Tang , W and Eisenbrand , C . 1992 . Chinese drugs of plant origin , Berlin : Springer . p. 983–988
- Vos , P , Hogers , R , Bleeker , M , Reijans , M , Van De Lee , T , Hornes , M , Frijters , A , Pot , J , Peleman , J , Kuiper , M and Zabeau , M . 1995 . AFLP: a new technique for DNA fingerprinting . Nucl Acid Res , 23 : 4407 – 4414 .
- Wu , JG , Li , ZY , Liu , Y , Liu , HL and Fu , TD . 1997 . Cytogenetics and morphology of the pentaploid hybrid between Brassica napus and Orychophragmus violaceus and its progeny . Plant Breed , 116 : 251 – 257 .
- Yang , L , Wu , SH , Zhang , QH , Liu , FY and Wu , P . 2007 . 23, 24-dihydrocucurbitacin B induces G2/M cell-cycle arrest and mitochondria-dependent apoptosis in human breast cancer cells (Bcap37) . Cancer Lett , 256 : 267 – 278 .