Abstract
One of the major topics in primate evolution is the phylogenetic position of the bizarre Daubentonia madagascariensis (DMA, aye-aye). The principal points that have been discussed for many decades are whether the aye-aye is: (i) the sister group of primates; (ii) the sister group of strepsirhines; or (iii) the sister group of lemurs. Very little is known about Daubentonia evolution, particularly on the chromosomal background. The present report focuses on the chromosomal history of this species. We used available chromosome painting data as the main source to identify conserved chromosomes, chromosomal segments and syntenic associations that have characterized the aye-aye karyotype. The dataset includes 47 characters that have been subjected to a concatenated analysis using maximum parsimony (MP) and Bayesian inference (BI). Both MP and BI topologies show Daubentonia as an independent monophyletic lineage, sister group of all other Strepsirhini. Further, both trees have weak statistical support as result of the high number of autapomorphies and homoplasies that have characterized the history of this group of species.
Introduction
The Strepsirhini(or Prosimii) represents one of the two major primate groups. It can be divided into three infraorders: the Chiromyformes (with only one representative: Daubentonia madagascariensis (DMA), the aye aye), the Malagasy Lemuriformes and the African and Asian Lorisiformes (Groves Citation2001). The Feagle (Citation1998) taxonomy, and more recent taxonomic interpretation, separated Lemuriformes into five families: Cheirogaleidae, Lemuridae, Lepilemuridae, Indriidae, and Daubentoniidae (Mittermeier et al. Citation2008). Phylogenetic relationships between these taxa are enigmatic (i.e., Roos et al. Citation2004; Froenicke Citation2005; Murphy et al. Citation2005; Horvarth et al. Citation2008).
Our interest focuses on the Daubentoniidae. The family has a sole living species, Daubentonia madagascariensis, the aye-aye, which is the world’s largest nocturnal primate (Figure a, b). It has a very controversial systematic and phylogenetic position (Dene et al. 1976; Schwartz and Tattersall Citation1985; Rumpler et al. Citation1988; Yoder et al. Citation1996; Groves Citation2004). Several morphological features place D. madagascariensis as the ancestral representative of the Malagasy radiation, with a basal position in the phylogeny of the group.
Figure 1 (a) Daubentonia madagascariensis in a sketch from Jophen Wolf (1832–1899) and (b) a skull from the Frans Lanting photo collection (modified).
Conversely many cranial, mandibular and dental features have located Daubentonia in a clade together with the Indriidae (a round skull and thicker lower incisor) (Schwartz and Tattersall Citation1985).
Several molecular phylogenies propose two main ramifications: that Daubentonia (i) is a sister taxon of the other Malagasy lemurs (DelPero et al. Citation1995; Yoder et al. Citation1996; Yoder Citation1997; Pastorini et al. Citation2002; Go et al. Citation2005); or (ii) is a primitive stock to all other prosimians (e.g. Adkins and Honeycutt Citation1994; Arnason et al. Citation1998). Nevertheless, molecular approaches do not shown a sufficient power of resolution for this species.
Karyological data did not support any of previous hypotheses. DMA displays a rearranged karyotype, symptomatic of a decided drift from the ancestral genome (Rumpler et al. Citation1988; Rakotoarisoa et al. 2000). A similar condition is present in all lemuriforms, which are characterized by a high rate of chromosomal dynamics and genome reshuffling. At the present the unique cytogenetic evidence for this genus highlights how Daubentonia shares some very ancient chromosomal arrangements with non-primate mammals (Warter et al. Citation2005).
The present study attempts to provide a phylogenetic interpretation of this genus using chromosomal features as sources in order to improve previous hypotheses concerning its critical position. Chromosome painting data of DMA have been compared with homologous data from representatives of prosimians and Scandentia. This allows us to define an interpretative tree of syntenic associations, with the scope of finding a phylogenetic meaning under cladistic constrains.
Materials and methods
We collect all published chromosome painting data from eight species within lemuriformes (Daubentonia madagascariensis, Lemur catta, Eulemur fulvus, Microcebus murinus, Avahi laniger, Indri indri, Lepilemur mustelinus, and Lepilemur ruficaudatus) and three species within lorisiformes (Galago moholi, Otolemur crassicaudatus, and Nycticebus coucang). Tupaia belangeri (Scandentia) was included as outgroup.
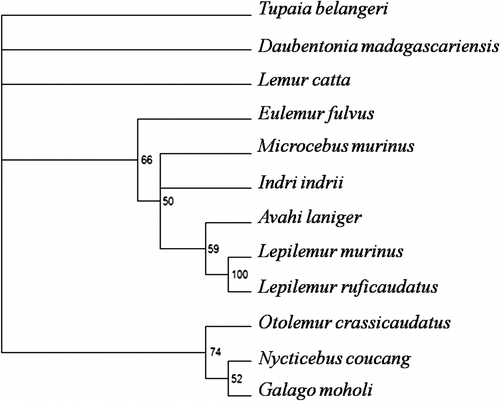
The data were subjected to an unweighted maximum parsimony (MP) analysis (PAUP 4.0, Swofford 2002), using a heuristic search with 100 random sequence additions and (tree-bisection-reconnection branch swapping) TBRbranch swapping (Figure ).
Figure 2 Strict consensus of the eight most parsimonious trees (tree length = 58, CI = 0.53, RI = 0.72) obtained from the maximum parsimony analysis. Numbers at the nodes indicate the statistical support values obtained from 1000 bootstrap replicates (only shown if > 50%).
Robustness of the inferred trees was explored through consistency indexes (CI) and retention indexes (RI), and support for each node was assessed using 2000 bootstrap replicates. A Bayesian inference was then performed using MrBayes version 3.2 (Ronquist and Huelsenbeck Citation2003) (Figure ). A total of 47 characters were identified based on the presence/absence of human segmental associations in the Strepsirhini and outgroup taxa, and coded as shown in the data matrix (Table )
Figure 3 Most likely phylogenetic reconstruction derived from the Bayesian analysis. Numbers adjacent to the nodes indicate estimated posterior probabilities.
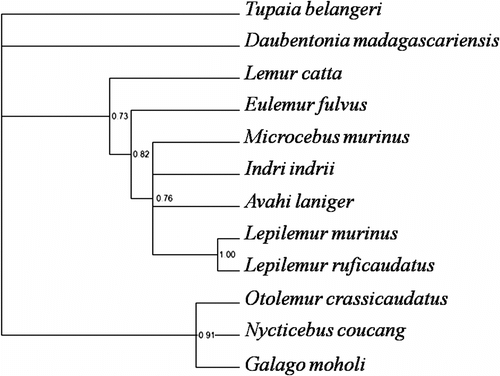
Table 1. The data matrix subjected to PAUP. Two codes have been established: 0 = absent; code 1 = presence of shared human syntenic associations.
Results and discussion
Eight equally parsimonious trees (L = 58, CI = 0.53, RI = 0.72 excluding non-informative characters) were obtained by MP analysis, and the strict consensus of these is shown in Figure .
An additional analysis has been performed using the Bayesian method (posterior probabilities are shown at the branches) (Figure ). Our comparison shows interesting results. Both topologies are only weakly supported both by bootstrap values at most of the nodes (MP tree) and Bayesian support values. This probably reflects the high numbers of autapomorphies and homoplasies that have accumulated during the chromosomal evolution of this lineage (HSA4/9, 7/12/22, 7/13, 10/13, 17/7, 18/7, 19/20). Invariably, in both reconstructions Daubentonia is a monophyletic lineage, thus rendering this species unique and independent of the lemuriform group. This result has been confirmed by the recent genome sequence of the aye-aye (Perry et al. Citation2011).
Additionally, the aye-aye maintains most of the ancestral derived associations that have been postulated for the AEK, and that characterized many mammalian species (HSA3/21, 14/15, 7/16, 12/22 (twice), and 16/19) but not all prosimians (Table ). Table highlights another interesting aspect that underlines the distinctiveness of Daubentonia: the presence of the HSA7/12/22 syntenic association, which represents a cytogenetic signature for squirrels (Li et al. Citation2004) (Rodentia). Further a HSA7/12 association is also found in all lorisiformes studied by a chromosomal painting approach. This is a supplementary result that may confirm the early split of Daubentonia genus from the other prosimians’ lineage.
Table 2. Comparison between Daubentonia madagascariensis and other prosimians based on human syntenic associations established by chromosome painting using human as probe. “A” indicates ancestral shared characters; Light gray bars indicate derived characters shared by Lemuriformes; dark gray bars indicate derived characters shared by Lorisiformes. Black bars are autapomorphies for D. madagascariensis
Comparative chromosome studies have then confirmed the diversity of Daubentonia genome.
The diploid number is 2n = 30 chromosomes, one of the lowest numbers of chromosomes among primates. The karyotype consists of seven pairs of metacentrics, five of submetacentrics, and two pairs of acrocentrics. The X and Y chromosomes are metacentric (Rumpler et al. Citation1988; Poormann-Allen and Izard 1990). Contrary to the abundant contributions attributed to morphological or molecular studies, chromosomal reports are very few, diversely to the conspicuous contribute produced by morphological or molecular studies, chromosomal reports are very decant (Rumpler et al. 1988).
Table shows a list of the human syntenic associations that have been maintained in Daubentonia and in other lemuriform genomes.
Chromosome painting studies detected ancestral simplesiomorphies and synapomorphies as syntenic blocks shared among different lineages. According to these reconstructions a diploid number of 46 has been postulated for the ancestral Eutherian karyotype (AEK) (Froenicke et al. 2005), which includes 10 human chromosomes (1, 5, 6, 9, 11, 13, 17, 18, 20 and X) conserved in toto, and seven syntenic associations formed from two or more human chromosomes (3p/21, 4/8p, 10p/12q/22qt, 12qt/22q, 14/15, 16p/7a and 19q/16q). In this scenario chromosome painting data proved that strepsirhine karyotypes are as derived as other primate genera (Warter et al. Citation2005; Roberto et al. Citation2008). All prosimian genomes are linked by a simplesiomorphic share condition that had evolved into a more derived status in the lemuriformes, producing the present chromosomal organization.
Cytogenetic studies have demonstrated a lack of derived syntenic associations shared by Lorisiformes and Lemuriformes. These include HSA2/4, 4/6 and 8/15 associations in all lemuriforms (Carbone et al. Citation2002; Warter et al. Citation2005), and HSA1q/19p, 2/12, 6/14, 12/7/16, 9/15 and 10/19q association in lorisiforms (Stanyon et al. Citation2002; Nie et al. Citation2005), with the exception of Daubentonia madagascariensis which certainly occupies an uncharacteristic position (present study). This has already been observed by early chromosomal banding studies (R-banding) where Daubentonia unequivocally held a distinct position from other lemurs (Rumpler et al. Citation1988; Poormann-Allen and Izard 1990; Dutrillaux and Rumpler 1995).
In conclusion the position of the aye-aye is still one of the major problems in Strepsirhini phylogeny. In fact, Daubentonia has an autapomorphic nature in many aspects.
| 1. | Body proportion and skeleton measurements indicate that Daubentonia is strictly different from other primates. An evolutionary and adaptive evaluation of its morphology supports Daubentonia within the order but does not support a strict relationship with all other Strepsirhini, leading to the hypothesis of a sister-group ranking. | ||||
| 2. | Molecular studies place Daubentonia as a first divergent taxon with two possible consequences. being an ancestor or alternatively a sister group to other Malagasy lemurs. | ||||
| 3. | Chromosomal analysis has not resolved the controversial position of Daubentonia. Our study neither proves nor disproves the staus of Daubentonia within the context of its position in the primate tree. | ||||
In conclusion, the current work is an in silico study of chromosomal data from previous publications on the chromosomal evolution of the aye-aye of Madagascar (Daubentonia madagascariensis). Our goal is to determine how the phylogenetic position of this species diverges when chromosomal features have been used as input source and mapped into a phylogenetic framework.
In this context, a reconstruction based on chromosomal syntenic groups revealed Daubentonia to contain several chromosomal characters that are not shared with other primates (lemurs, lorises, or anthropoids), but are shared with non-primate mammalian relatives. This raises serious questions about the nature of karyotypic change.
Acknowledgments
BP is a postdoctoral fellow at SANBI, University of Western Cape. She is grateful to colleagues for support and advice. BP and LS thank Professor Takafumi Ishida for critical reading of the manuscript.
References
- Adkins , RM and Honeycutt , RL . 1994 . Evolution of primate cytocrome c oxidase subunit II gene . J Mol Evol , 38 ( 3 ) : 215 – 231 .
- Arnason , U , Gullberg , A and Janke , A . 1998 . Molecular timing of primate divergences as estimated by two nonprimate calibration points . J Mol Evol , 47 ( 6 ) : 718 – 727 .
- Carbone , L , Ventura , M , Tempesta , S , Rocchi , M and Archidiacono , N . 2002 . Evolutionary history of chromosome 10 in primates . Chromosoma , 111 ( 4 ) : 267 – 272 .
- DelPero , M , Crovella , S , Cervella , P , Ardito , G and Rumpler , Y . 1995 . Phylogenetic relationships among Malagasy lemurs as revealed by mitochondrial DNA sequence analysis . Primates , 36 ( 3 ) : 431 – 440 .
- Dene H, Goodman M, Prychdko W, Moore GW. 1976. Immunodiffusion systematics of the primates. Folia Primatol. 25(1):35–61.
- Dutrillaux B, Rumpler Y. 1995. Phylogenetic relations among prosimii with special reference to Lemuriformes and Malagasy nocturnals. In: Alterman L, Azard MK, editors. Creatures of the dark: the nocturnal prosimians, New York (NY): Plenum Press. p. 141–150.
- Feagle , JG . 1998 . Primate adaptation and evolution , New York : Academic Press .
- Froenicke , L . 2005 . Origins of primate chromosomes—as delineated by Zoo-FISH and alignments of human and mouse draft genome sequences . Cytogenet Genome Res , 108 ( 1–3 ) : 122 – 138 .
- Go , Y , Rakotoarisoa , G , Kawamoto , Y , Shima , T , Koyama , N , Randrianjafy , A , Mora , R and Hirai , H . 2005 . Characterization and evolution of major histocompatibility complex class II in the aye-aye, Daubentonia madagascariensis . Primates , 46 ( 2 ) : 135 – 139 .
- Groves , CP . 2001 . Primate taxonomy , Washington (DC) : Smithsonian Institution Press .
- Groves , CP . 2004 . “ Charles Oxnard and the aye-aye: morphometrics, cladistics, and two very special primates ” . In Shaping primate evolution: form, function and behavior , Edited by: Anapol , F , German , RZ and Jablonski , NG . 368 – 377 . Cambridge (UK) : Cambridge University Press .
- Horvarth , J , Weisrock , DW , Embry , SL , Fiorentino , I , Balhoff , JP , Kappeler , P , Wray , GA , Willard , HF and Yoder , AD . 2008 . Development and application of a phylogenomic toolkit: resolving the evolutionary history of Madagascar’s lemurs . Genome Res , 18 ( 3 ) : 489 – 499 .
- Li , T , O’Brien , PC , Biltueva , L , Fu , B , Wang , J , Nie , W , Ferguson-Smith , MA , Graphodatsky , AS and Yang , F . 2004 . Evolution of genome organizations of squirrels (Sciuridae) revealed by cross species chromosome painting . Chromosome Res , 12 ( 4 ) : 317 – 333 .
- Mittermeier , RA , Ganzhorn , JU , Konstant , WR , Glander , K , Tattersall , I , Groves , CP , Rylands , AB , Hapke , A , Ratsimbanzafi , J , Mayor , MI , Loius , EE Jr , Rumpler , Y , Schwitzer , C and Rasoloarison , RM . 2008 . Lemur diversity in Madagascar (PDF) . Int J Primatol , 29 ( 6 ) : 1607 – 1656 .
- Murphy , WJ , Larkin , DM , Everts-van der Wind , A , Bourque , G , Tesler , G , Auvil , L , Beever , JE , Chowdhary , BP , Galibert , F , Gatze , L , Hitte , C , Meyers , SN , Milan , D , Ostrander , EA , Pape , G , Parker , HG , Raudsepp , T , Rogatcheva , MB , Schook , LB , Skow , LC , Welge , M , Womack , JE , O'Brien , SJ , Pevzner , PA and Lewin , HA . 2005 . Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps . Science , 309 ( 5734 ) : 613 – 617 .
- Nie , W , O’Brien , PCM , Wang , J , Su , W , Robinson , TJ and Yang , F . 2005 . Chromosome painting between human and lorisiform prosimians: evidence for the HSA 7/16 synteny in the primate ancestral karyotype . Am J Physical Anthropol , 129 ( 2 ) : 250 – 259 .
- Pastorini , J , Forstner , MRJ and Martin , RD . 2002 . Phylogenetic relationships among Lemuridae (Primates): evidence from mtDNA . J Human Evol , 43 ( 4 ) : 463 – 478 .
- Perry GH, Reeves D, Melsted P, Ratan A, Miller W, Michelini K, Louis EE, Pritchard KJ, Mason EC, Gilad E. 2011. A genome sequence resource for the aye-aye (Daubentonia madagascariensis), a nocturnal lemur from Madagascar. Genome Biol Evol. 4(2):126–135 doi:10.1093/gbe/evr132
- Poorman-Allen , P and Izard , MK . 1990 . Daubentonia madagascariensis (Primates, Daubentoniidae) . Int J Primatol , 11 ( 5 ) : 401 – 410 .
- Ratokoarisoa , G , Hirai , Y , Go , Y , Kawamoto , Y , Shima , T , Koyama , N , Randrianjafy , A , Mora , R and Hirai , H . 2000 . Chromosomal localization of 18S rDNA and telomere sequence in the aye-aye, Daubentonia madagascariensis . Genes Genet Syst , 75 ( 5 ) : 299 – 303 .
- Roberto , R , Capozzi , O , Wilson , RK , Mardis , ER , Lomiento , M , Tuzun , E , Cheng , Z , Mootnick , AR , Archidiacono , N Rocchi , M . 2008 . Molecular refinement of gibbon genome rearrangements . Genome Res , 17 ( 2 ) : 249 – 257 .
- Ronquist , F and Huelsenbeck , JP . 2003 . MrBayes 3: Bayesian phylogenetic inference under mixed models . Bioinformatics , 19 ( 12 ) : 1572 – 1574 .
- Roos , C , Schmitz , J and Zischler , H . 2004 . Primate jumping genes elucidate strepsirrhine phylogeny . PNAS , 101 ( 24 ) : 10650 – 10654 .
- Rumpler , Y , Warter , S , Petter , JJ , Albignac , R and Dutrillaux , B . 1988 . Chromosomal evolution of Malagasy lemurs. XI. Phylogenetic position of Daubentonia madagascariensis . Folia Primatol , 50 ( 1–2 ) : 124 – 129 .
- Schwartz , JH and Tattersall , I . 1985 . Evolutionary relationships of living lemurs and lorises (Mammalia, Primates) and their potential affinities with European Eocene Adapidae . Anthropol Pap Am Mus , 60 : 1 – 100 .
- Schulze-Hagen K. and Geus A. 2000. Joseph Wolf (1820-1899): Tiermaler = Joseph Wolf (1820-1899): animal painter. Basilisken-Presse: Marburg an der Lahn. 361pp.
- Stanyon , R , Koehler , U and Consigliere , S . 2002 . Chromosome painting reveals that galagos have highly derived karyotypes . Am J Phys Anthropol , 117 ( 4 ) : 319 – 326 .
- Swofford DL. 2002. PAUP∗: Phylogenetic analysis using parsimony (∗and other methods) v. 4.0b10. Sunderland, MA: Sinauer Associates.
- Warter , S , Hauwy , M , Dutrillaux , B and Rumpler , Y . 2005 . Application of molecular cytogenetics for chromosomal evolution of the Lemuriformes (Prosimians) . Cytogen Genome Res , 108 ( 1–3 ) : 197 – 203 .
- Yoder , AD . 1997 . Back to the future: a synthesis of strepsirhine systematic . Evol Anthropol , 6 ( 4 ) : 11 – 22 .
- Yoder , AD , Cartmill , M , Ruvolo , M , Smith , K and Vilgays , R . 1996 . Ancient single origin for Malagasy primates . Proc Natl Acad Sci USA , 93 ( 10 ) : 5122 – 5126 .