Abstract
The current work is an in silico study of data from previous publications and genome browsers, on the origin of the human synteny HSA7a/16p. The molecular composition of the chromosomal segments identified as HSA7a/16 and 7b (free or differently associated) is not yet clear. This means that a syntenic association 7/16, which can be detected by an in situ hybridization (FISH) method in different taxa, may not necessarily correspond to those of the same association in different lineages. In silico research, together with comparative cytogenetics, have been applied in order to define the composition of the 7/16 syntenic association. These results allow a confident reconstruction of the syntenic associations HSA7/16 in diverse vertebrate lineages, indicating various levels of homology, but also considerable levels of non-homology, which should elicit caution with phylogenetic interpretations. The 7/16 association represents a paradigmatic example of the complexity involved in the interpretation of comparative cytogenetic data from a phylogenetic perspective.
Introduction
The development of powerful molecular cytogenetic and genomic technologies such as Zoo-FISH (Zoo-fluorescence in situ hybridization) has facilitated large-scale genome comparisons for a different number of species. Based between the hybridization of labeled DNA probes and genomic DNA, in situ hybridization techniques allow for the unequivocal confirmation of homology among chromosomes. Additionally an even more precise assessment of structural rearrangement is provided by the use of the reciprocal cross-species painting schemes (Yang et al. Citation2000; Weinberg 2005). Data from chromosomal painting allow the reconstruction of “ancestral karyotypes”. Many comparisons have been made with the human genome so that conserved syntenic blocks defined by homology with human chromosomes can be recognized for all species. Although this approach is useful for identifying ancestral chromosome states, unless such data are placed in a correct cladistic framework, they do not necessarily provide nodal support for specific cladogenetic events. The need to discriminate shared ancestral simplesiomorphies from derived ancestral synapomorphies generates the challenge of resolving nodes in eutherian phylogenetic trees using Zoo-FISH. In this context the Human Genome Project (HGP) has built a reasonable challenge to the comprehension has built a reasonable challenge to the understanding of the architecture and the evolution of the human genome of the architecture and the evolution of the human genome (Collins et al. Citation2003; Hardison et al. Citation2003; Weise et al. 2005). Therefore, with the advent of full genome assemblies for several vertebrates, including the opossum, the chicken and some fishes, it is now possible to reinterpret chromosomal characters to better define the ancestral condition (www.genome.ucsc.edu; www.ensembl.org). Currently 41 mammalian species have been partially or fully sequenced, including: 18 species of Euarchontoglires, 13 of Laurasiatheria, three of Afrotherian, two of Xenarthra, three of Metatheria, and one of Prototheria. To date only nine genomes have been completed; this allows screening of syntenic blocks in pairwise genome comparison. These resources prominently enriched the efficacy of the genome comparative analyses, and provide a useful tool for better understanding chromosomal architectures. Our database includes sources for the number of species available when the analysis was performed.
The starting point of the present analysis is human chromosome 7, which in its extant composition is the result of a fusion between ancestral elements identified as 7a and 7b (Muller et al. 2004; Romagno et al. Citation2004). From chromosome painting and reciprocal painting on several living taxa, and from the reconstruction of the ancestral eutherian karyotype, we know that fragments of 7 and 16p are syntenic, but the contribution of 7 in this association is not clear. However, it transpires that this association is present in different lines of vertebrates and, given its intrinsic syntenic composition, assumes importance as a marker of homology or homoplasy in phylogenetic reconstructions.
The present work investigates the composition of the 7a fragment and of the entire 7 association, as a result of doubt regarding the consistency of the composition of segment 7, in the 7/16 association, in different evolutionary lines of the subphylum. A combination of molecular cytogenetics and computational approaches has contributed to our understanding of the origin of the evolution of the syntenic association 7/16, and to the identification of specifically conserved genomic regions. Moreover, these findings are of paradigmatic significance, highlighting the need for caution in the interpretation of painting data and in their application to solving phylogenetic questions.
Materials and methods
Two different data sources were used to identify homologous synteny blocks (HSBs) and evolutionary breakpoints (EBs) between the genomes of humans and various mammalian species. The species analyzed are presented in Table . We proceed as follows: as a first step we collected and analyzed chromosome-painting data from 36 mammalian species. Subsequently, we devised phylogenetic hypotheses to delineate the dynamics of HSA7/16 syntenic association.
Table 1. List of species analyzed by reciprocal chromosome painting using human paints and/or the locus specific probe WS (HSA7q11.23) and/or comparisons of whole genome assemblies. The numbers of human chromosomes 7 and 16 orthologous blocks (OB) are indicated for each species. RP (reciprocal painting); WGA (whole genome amplification); BAC (bacterial artificial chromosome); PAC (plasmid artificial chromosome); CP (chromosome painting); WS (William’s Beuren single mapping); RH (radiation hybrid mapping)
The second step involved focusing on 15 species for which reciprocal chromosome painting data, whole genomic comparison (Ensembl database) and the original locus specific mapping of probe WS (7q11.23) were available. In the third step multiple comparisons were considered crucial for the reconstruction of phylogenetic trees for the human syntenic associations 7/16 in various vertebrate lineages (Figure ). In the fourth step, on the basis of the reciprocal painting and sequence data (www.ensembl.org) collected, we were able to construct a summary table of breakpoints. This allowed us to detect the human chromosome 7 and 16 band composition in the different orders (Supplementary Table available via the article website).
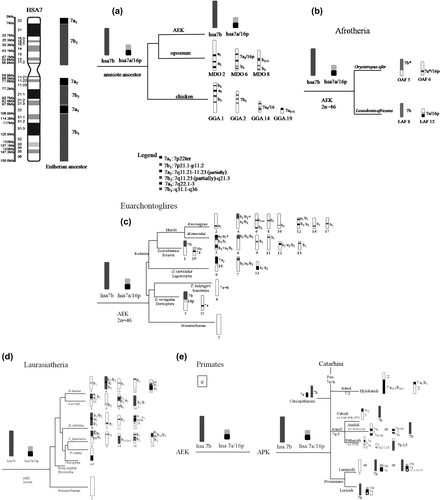
Figure 1 Fifteen species assigned to 12 orders were included in the sample to track the evolution of the ancestral synteny HSA7/16 using phylogenetic reconstruction. The ideogram of the HSA7 has been extracted from Ruiz-Herrera et al. (Citation2006). The choice of species was influenced by the availability of reciprocal painting data, whole genome comparisons and locus specific probe WS (7q11.23). Mammalian trees include: (a) Aves (chicken) and Therian (opossum); (b) Afrotheria; (c) Euarchontoglires; (d) Laurasiatheria; and (e) Primates (Strepsirhini and Haplorhini). Black segments are homologs of HSA7a, dark gray segments are homologs of HSA7b, whereas light gray segments are homologs of HSA16p.
Results and discussion
This section is divided into two parts. The first part describes the results of the organization of the chromosome bands in mammals, with a reconstruction of: (i) their evolution; (ii) the constitution of extant human chromosome 7; (iii) the architecture of the syntenic association; and (iv) the new description of 7a (a1, 2, 3) and 7b (b1, 2, 3). Landmarks were obtained via a comparative study of data from all available sources. Analysis of the genome assemblies of mammalian genomes can help to test hypotheses about the content of the ancestral eutherian genome as previously defined by the molecular cytogenetics approach. The second part, while adopting the same approach, focuses solely on non-human primates as a model, given their similarity to the human genome.
Our combined analysis has defined the complex composition of the 7a and 7b fragments, with the identification of three fractions, respectively, as follows: [7a1(7pt22-7pter); 7b1 (7p21.1-7p11.2); 7a2 (7q11.21-7q11.23, partially); 7b2 (7q11.23, partially-7q21.3); 7a3 (7q22.1-7q3.1); and 7b3 (7q31.1-7q3.6)].
The “a” and “b” distinctions, although potentially misleading, have been maintained in order to facilitate the comprehension of comments on previous descriptions derived from the literature.
HSA7/16 ancestral architecture in mammals and the history of evolutionary breakpoints
Using human chromosome 7 as reference (the ideogram extracted from Ruiz-Herrera et al. [2006] has been included in Figure ), homologous syntenic blocks have been identified for different species of eutherian mammals (representative of the recognized genomic macro groups: Afrotheria, Eurchontoglires, Laurasiatheria), with the addition of one Methatherian (marsupial) and one member of the class Aves (Figure ) which have been included in the out-group, as potentially “ancestral”.
Four phylogenetic trees are proposed, to summarize the evolution of the ancestral chromosome 7 in these species. The first (Figure a), takes into account the genome organization in Aves (Gallus gallus, GGA) and Metatherian (Monodelphis domestica-MDO) (see also Robinson & Ruiz-Herrera Citation2008). M. domestica (chromosome 6-MDO6) and G. gallus (chromosome 14) are characterized by the HSA7a1 /16, where 7a1 has a 7p22-pter composition. This analysis also highlights another 7a chromosomal band on different chromosomes (see GGA1 and MDO2, Figure a). Both lineages have maintained the same HSA7a1 /16p configuration.
The 7b fragment appears to be divided into two fragments in both species in this group. The chromosomal band 7b1 is partially linked to HSA7a1/16 on MDO6, and the rest of 7b1 is distributed on the MDO8 chromosome (associated with b2+3). In the chicken genome, 7b1 is mainly located on the GGA2 chromosome, and a small portion is associated with another synteny on GGA1 (Figure a).
The first node in the ancestral eutherian karyotype may be represented by the Afrotherians (i.e. Orycteropus afer and Loxodonta Africana) (Figure b). Chromosome 6 of Orycteropus afer (OAF, Tubulidentata) and chromosome 12 of Loxodonta Africana (LAF, Proboscidea) show the whole ancestral association HSA7a/16p (7p22ter/7q11.21-11.23, partially/7q22.1-3/16p) and an uninterrupted orthologous 7b (Figure b). Interestingly, a different result has been proposed by Robinson et al. (2004) which proposes 7b/16p as ancestral synteny association. This represents an important contradiction to the previous supposed ancestral synteny, the 7b/16p association, proposed by Robinson et al. (Citation2004).
For Euarchontoglires, a third scenario is proposed (Figure c). Mouse and rat are characterized by the highest number of chromosomes breakpoints that involve the ancestral synteny, leading to the presence of four and three pieces of 7a fragments respectively, and eight/10 pieces of the 7b fragment in mouse and rat (see Supplementary Table available via the article website). Therefore, in Rodentia there is no trace of the ancestral eutherian 7a/16p association.
In Galeopterus variegatus (GVA), Dermoptera, the orthologous to HSA7 is divided into two parts. The 7/16p synteny is part of GVA chromosome 3. It is proposed here that this association involves the 7b segment (see also Nie et al. Citation2008). On this basis, Sciuridi (S. carolinensis) and Dermoptera (G. variegatus) are phylogenetically divided by the organization of the 7/16 association. This is a crucial point in the eutherian ancestral karyotype hypothesis.
In Lagomorpha, the 7/16 association is present but with a different composition. A 7a2/16p is present on Oryctolagus cuniculus (OCU) chromosome 6. Evolutionary breakpoints at 7p21 and 7q21.1 are responsible for the fragmentation of both ancestral segments 7a and 7b, into four and two pieces, respectively.
In Scandentia (Tupaia belangieri, TBE), there is no trace of the ancestral 7/16 association, and the HSA7 configuration has been retained as an intact orthologous block (see also Müller et al. 1999).
Significant numbers of intra-chromosomal rearrangements are at the base of the Laurasiatheria clade diversification (Figure d).
Artiodactyla is the most diverse order. Chromosome 7 segments are differently associated: 7b1 (7p21.1-p11.2)/16p (i.e. Bos taurus, chromosome 25) and the HSA7a3 (7q22.1-3)/16p (i.e. Sus scrofa, chromosome 3) were the syntenic associations found.
In Perissodactyla (Equus caballus, EQU), the ancestral syntenic association involves HSA7a2 (7p22ter) and HSA7a3 (7q22.1-3) on the EQU13.
In Carnivora, Canis familiaris (CFA), HSA7b2 /16p localizes on CFA 6 (HSA7q11.23-q21.3). Other pieces are highly fragmented and distributed on different chromosomes (CFA6, 9, 14, 16 and 18). In the cat (Felis cattus, FCA) there is no evidence of the HSA7/16 synteny.
HSA7/16 chromosomal organization in primates
The order of primates shows a complex evolutionary scenario (Figure e). Within Strepsirrhini (Prosimians), lorisiformes (Galagidae and Lorisidae) are characterized by a single block containing only sequences orthologous to HSA7b. Another chromosome maintains the 7a/16p ancestral association, normally associated with the human chromosome 12. Evidence was found that HSA7p21, HSA7q21.1 and HSA7q31.1 are the breakpoints involved in the 7/16 association in prosimians.
Lemuriformes genera are characterized by several associations, which have led to vivacious organizations of the ancestral HSA7a/16p: i) a HSA7a/16p association and 7b segment (i.e., Eulemur, Avahi, Propithecus); ii) a HSA7a/16p association involved only with other human syntenies (i.e., HSA1, 12, 17), and founded in Daubentonia genus; and iii) the entire synteny HSA7 (7a+7b) completely conserved in Lemur catta.
In Platyrrhini there is a 7a/5 tandem translocation, which is unique to this lineage, and which has led to three different patterns of evolution: 7a/5 (7q11.23/5) in Cebidi, 7a/5/7a (7q11.21-11.23/5/7p22ter+7q22.1-3) in Atelidi, and 7a/5/7a/5 in Pithecidi. The segment 16p is associated to the human synteny HSA10. Our analysis of the reciprocal chromosome painting on Lagothrix lagothrica (Atelidi) indicates the occurrence of two breakpoints (HSA7p15.3 and HSA7q22.1) in the HSA7/5 association.
Catarrhini are characterized by a genomic enrichment by duplications and different inversions (Sineo et al. Citation2007; Picone et al. Citation2008), showing that reconstruction of ancestrals can be complicated. Synteny 7 is entire in the great majority of species, but our reconstruction brings to light some interesting results. Namely, Hominidae (Pongo, Pan and Gorilla) and Cercopithecidae (Macaca) share a breakpoint at HSA7p13, and two evolutionary breakpoints at HSA16p13.1 and 16p22. Also, a pericentric inversion, involving three breakpoints at HSA7p13, HSA7p22 and HSA7q11.23, is responsible for the difference between Pongo pygmaeus (PPY), Gorilla gorilla (GGO) and humans, whereas we discovered HSA7p13and HSA7p22 to be the evolutionary breakpoints in Pan troglodytes (PTR).
Moreover, among Cercopithecidae, Macaca mulatta (MMU) displays the largest fraction of breakpoints (HSA7p13, HSA7p15.3, and HSA7p15.2). Conversely, Chlorocebus aethiops (CAE) is the only species in which HSA7 is present as two distinct orthologous blocks: HSA7a on CAE 28 and HSA7b on CAE21. Finally, Hylobatidae karyotypes (lesser apes) appear to be highly derived and rearranged (see also Misceo et al. Citation2008). Also, the lesser ape genome shows different degrees of HSA7 disruption and association: HSA7a2+3/b2+3/2 and HSA7a1/b1/2.
Breakpoints analysis
Supplementary Table summarizes a list of evolutionary breakpoints in primates and other mammals. Data from comparative FISH analysis have been retrieved in order to resolve the evolutionary history of breakpoint regions for ancestral HSA7/16 synteny. Breakpoint regions were then analyzed by reference to the Ensembl database and the human genome as reference. In total, 143 orthologous blocks were found for human chromosome 7 and 89 for the human chromosome 16 in amniote genomes (Table ). As expected, the largest fraction of evolutionary breakpoints was found in the deepest branching (chicken and opossum) and the most highly rearranged in mouse and rat, and also in cattle and pig (Supplementary Table).
Muroid rodents have a large number of order specific breakpoints, whereas primates have the fewest. Thus, after the divergence between human and chimpanzee, the human lineage has had the largest fraction of evolutionary breakpoints in the deepest branching, compared to that of the chimpanzee.
Conclusions
The analysis puts forward a scenario from which it is possible to establish a diverse organization of the 7/16 syntenic association from its ancestral state. The study provides evidence of organizations in three superordinal clusters: (i) the Afrotheria, where the 7a/16p association is conserved and the 7a is composed of (7p22ter/7q11.21-11.23, partially/7q22.1-3/16p) (Figure a); (ii) the Euarchontoglires, where the chromosomal band HSA7p21 represents a reuse breakpoint for different orders, except Dermoptera – the entire HSA7a (7p22ter/7q11.21-11.23, partially/7q22.1-3) segment and HSA7q11.21-11.23 appear to be the boundaries of the 7/16 mammalian ancestral association (Figure b); (iii) Laurasiatheria is the most complex clade. Here the bands ranging from 7p21.1-p11.2 to 7q22.1-3 are the boundaries of the ancestral 7/16 association. Nevertheless this composition is not ascribed exactly to 7a or to 7b (Figure c).
The phylogenetic pattern recovered from our analysis indicates the presence of various breakpoints (Supplementary Table). Thus, HSA7q11.23 occurred in three different lineages: Eurachontoglires (Pongo pygmaeus, Gorilla gorilla, Galeopterus variegatus, Mus musculus, Rattus novergicus); Laurasiatheria (Bos taurus, Sus scrofa, and Equus caballus); and Galliformes (Gallus gallus). Similarly, the HSA16q22 is an evolutionary breakpoint for three different lineages: Euarchontoglires (Pongo pygmaeus, Gorilla gorilla, Pan troglodytes, Mus musculus, Rattus novergicus); Laurasiatheria (Bos taurus and Sus scrofa); and Galliformes (Gallus gallus).
The chromosomal bands extending from HSA7p15.2 to HSA7p11.2 have been maintained by Tupaia belangieri, Galago moholi, Nycticebus coucang, Lagothrix lagothrica and Loxodonta africana. Additionally, prosimians, rodents, Tupaia belangeri, Galago variegatus, Loxodonta africana, and Orycteropus afer are linked by sharing a large range of chromosomal bands (from 16q11.2 to 16q24) homologous to HSA16, and associated with other human syntenies.
Supplementary Table also indicates that band 7p13 houses nine evolutionary breakpoints and seven fragile sites; band 7q11.23 houses eight fragile sites and nine evolutionary breakpoints; and band 7q22.1 houses five fragile sites and six evolutionary breakpoints (Robinson et al. Citation2006; Ruiz-Herrera et al. Citation2006) (note that the information on number fragile sites has been extracted from Ruiz-Herrera et al. Citation2006).
This study has enabled us to examine how comparative molecular cytogenetic and computational approaches can contribute to the understanding of vertebrates’ genomic organization. Genomes have segments that have been conserved over hundreds of millions of years, and yet other regions, owing to their features, are unstable and more likely to be involved in rearrangements. The dimension of these regions is noteworthy, variable in size and association. It is important to consider how the increasing availability of fully sequenced genomes will alter current phylogenetic interpretations, and address previous imbalances, which resulted from the poor resolution of Zoo-FISH analyses on a limited number of species.
tcar_a_829691_sm2917.xls
Download MS Excel (38 KB)Acknowledgments
The authors would like to thank Susan Janse van Rensburg and colleagues in SANBI for their insightful comments and suggestions, and Claire Stypulkowsky for her help and comments during the writing. BP is a postdoctoral fellow supported by the South African National Research Foundation (NRF). Research has been supported by a grant from CORI-UNIPA (international collaboration projects) and by funds from University of Palermo, attributed to LS.
References
- Balmus G, et al. 2007. Cross-species chromosome painting among camel, cattle pig and human: further insights into the putative Cetartiodactyla ancestral karyotype. Chromosome Res. 15 (4): 499–514.
- Breen M, Thomas R, Binns MM, Carter NP, Langford CF. 1999. Reciprocal chromosome painting reveals detailed regions of conserved synteny between the karyotypes of the domestic dog (Canis familiaris) and human. Genomics. 61 (2): 145–155.
- Cardone MF, Ventur M, Tempesta S, Rocchi M, Archidiacono N. 2002. Analysis of chromosome conservation in Lemur catta studied by chromosome paints and BAC/PAC probes. Chromosoma. 111 (5): 267–272.
- Collins FS, Green ED, Guttmacher AE, Guyer MS. 2003. A vision for the future of genomics research. Nature. 422 (6934): 835–847.
- de Oliveira EHC, Neusser M, Figueiredo WB, et al. 2002. The phylogeny of howler monkeys (Alouatta, Platyrrhini): Reconstruction by multicolor cross-species chromosome painting. Chromosome Res. 10 (8): 669–683.
- Dumas F, Bigoni F, Stone G, Sineo L, Stanyon R. 2005. Mapping genomic rearrangements in titi monkeys by chromosome flow sorting and multidirectional in-situ hybridization. Chromosome Res. 13 (1): 85–96.
- Dumas F, Bigoni F, Stone G, Sineo L, Stanyon R. 2007. Phylogenomics of species from four genera of New World monkeys by flow sorting and reciprocal chromosome painting. BMC Evol Biol. 7 (2): 11–25.
- Hardison RC, Roskin K, Yang S, et al. 2003. Covariation in frequencies of substitution, deletion, transposition, and recombination during eutherian evolution. Genome Res. 13 (1): 13–26.
- Korstanje R, O’Brien PCM, Yang F, et al. 1999. Complete homology maps of the rabbit (Oryctolagus cuniculus) and human by reciprocal chromosome painting. Cytogenet Cell Genet. 86 (3–4): 317–322.
- Li T, O’Brien PCM, Biltueva L, et al. 2004. Evolution of genome organizations of squirrels (Sciuridae) revealed by cross-species chromosome painting. Chromosome Res. 12 (4): 317–335.
- Misceo D, Capozzi O, Roberto R, et al. 2008. Tracking the complex flow of chromosome rearrangements from the Hominoidea Ancestor to extant Hylobates and Nomascus Gibbons by high-resolution synteny mapping. Genome Res. 18 (9): 1530–1537.
- Müller S, Stanyon R, O’Brien PCM, Ferguson-Smith MA, Plesker R, Wienberg J. 1999. Defining the ancestral karyotype of all primates by multidirectional chromosome painting between tree shrews, lemurs and humans. Chromosoma. 108 : 393–400.
- Muller S, Finelli P, Neusser M, Wienberg J. 2004. The evolutionary history of human chromosome 7. Genomics. 84 : 458–467.
- Müller S, Stanyon R, Finelli P, et al. 2000. Molecular dissection of human chromosome 3 and 21 evolution. Proc Natl Acad Sci USA. 97 : 206–211.
- Murphy WJ, Davis B, David DA. 2007. A 1.5Mb-resolution radiation hybrid map of the cat genome and comparative analysis with the canine and human genome. Genomics. 89 (2): 189–196.
- Neusser M, Stanyon R, Bigoni F, Wienberg J, Muller S. 2001. Molecular cytotaxonomy of New World monkeys (Platyrrhini) a comparative analysis of five species by multi-color chromosome painting gives evidence for a classification of Callimico goeldii within the family of Callitrichidae. Cytogenet Cell Genet. 94 (3–4): 206–215.
- Nie W, Fu B, O'Brien PCM, et al. 2008. Flying lemurs – The 'flying tree shrews'? Molecular cytogenetic evidence for a Scandentia-Dermoptera sister clade. BMC Evol Biol. 1 (6): 18–29.
- Nie W, O’Brian PCM, Fu B. 2006. Chromosome painting between human and lorisiform prosimians: evidence for the HSA7/16 synteny in the primate ancestral karyotype. Am J Phys Anthrop. 129 (2): 250–259.
- Picone B, Dumas F, Stanyon R, et al. 2008. Exploring Evolution in Ceboidea (Platyrrhini, Primates) by Williams-Beuren probe (HSA 7q11.23) chromosome mapping. Folia Primatol. 79 (5): 417–427.
- Richard F, Lombard M, Dutrillaux B. 2000. Phylogenetic origin of human chromosomes 7, 16, and 19 and their homologs in placental mammals. Genome Res. 10 (5): 644–651.
- Robinson TJ, Fu B, Ferguson-Smith MA, Yang F. 2004. Cross-species chromosome painting in the golden mole and elephant shrew: support for the mammalian clades Afrotheria and Afroinsectiphillia but not Afroinsectivora. Proc Natl Acad Biol Sci USA. 271 (1547): 1477–1484.
- Robinson TJ, Ruiz-Herrera A. 2008. Defining the ancestral eutherian karyotype: a cladistic interpretation of chromosome painting and genome sequence assembly data. Chromosome Res. 16 (8): 1133–1141.
- Robinson TJ, Ruiz-Herrera A, Froenicke L. 2006. Dissecting the mammalian genome – new insights into chromosomal evolution. Trends Genet. 22 (6): 297–301.
- Romagno D, Chiarelli B, Sineo L. 2004. Evolution of human chromosome 7: new information from the mapping of Williams-Beuren locus on non human primates chromosome. Caryologia. 57 (1): 39–43.
- Ruiz-Herrera A, Castresana J, Robinson TJ. 2006. Is mammalian chromosomal evolution driven by regions of genome fragility? Genome Biol. 7 (12): 115–131.
- Sineo L, Dumas F, Vitturi R, Picone B, Privitera O, Stanyon R. 2007. Williams-Beuren mapping in Callithrix argentata, and Callicebus cupreus, and Alouatta caraya indicated different patterns of chromosomal rearrangements in Neotropical primates. J Zool Syst Evol Res. 45 (4): 366–371.
- Stanyon R, Bruening R, Stone G, Shearin A, Bigoni F. 2005. Reciprocal painting between humans, De Brazza’s and patas monkeys reveals a major bifurcation in the Cercopithecini phylogenetic tree. Cytogenet Genet Res. 108 (1–3): 175–182.
- Stanyon R, Consigliere S, Bigoni F, Ferguson-Smith M, O’Brien PC, Wienberg J. 2001. Reciprocal chromosome painting between a New World primate, the woolly monkey, and humans. Chromosome Res. 9 (2): 97–106.
- Stanyon R, Dumas F, Stone G, Bigoni F. 2006. Multidirectional chromosome painting reveals a remarkable syntenic homology between the greater galagos and the slow loris. Am J Primatol. 68 (4): 349–359.
- Trifonov VA, Stanyon R, Nesterenko AI. 2008. Multidirectional cross-species painting illuminates the history of karyotypic evolution in Perissodactyla. Chromosome Res. 16 : 89–107.
- Warter S, Hauwy M, Dutrillaux B, Rumpler Y. 2005. Application of molecular cytogenetics for chromosomal evolution of the Lemuriformes (Prosimians). Cytogenet Genome Res. 108 (1–3): 197–203.
- Weise A, Starke H, Mrasek K, Claussen U, Liehr T. 2005. New insights into the evolution of chromosome 1. Cytogenet Genome Res. 108 : 217–222.
- Wienberg J, Stanyon R, Nash WG. 1997. Conservation of human vs. feline genome organization revealed by reciprocal chromosome painting. Cytogenet Cell Genet. 77 (3–4): 211–217.
- Yang F, Alkalaeva EZ, Perelman PL. 2003. Reciprocal chromosome painting among human, aardvark, and elephant (superorder Afrotheria) reveals the likely eutherian ancestral karyotype. Proc Natl Acad Sci USA. 100 (3): 1062–1066.
- Yang F, Graphodatsky AS, O’Brien PCM. 2000. Reciprocal chromosome painting illuminates the history of genome evolution of the domestic cat, dog and human. Chromosome Res. 8 (5): 393–404.