Abstract
Seedless cultivars of banana are generally triploids, and diploids or tetraploids are rarer. In the present study, the karyomorphology of eight edible Musa cultivars of West Bengal was investigated for the first time to contribute to a better understanding of chromosome features and chromosome changes in the way of variation. Among the eight cultivars, seven were triploid and only the eighth seeded cultivar was diploid, all with basic chromosome number of x = 11. Karyotypic differences were observed between the cultivars but not enough to serve as a marker for each cultivar. Karyotype analysis has shown predominance of chromosomes with centromeres in median position with a few submedian centromeres. This report is the first of its kind for Musa cultivars where comparative karyotyping indicate that minor structural alterations of the chromosomes may have resulted in the genetic variability within the cultivated bananas, giving rise to numerous varieties or cultivars.
Keywords:
Introduction
The plant genus Musa is of extraordinary significance to human societies as it produces the fourth most important food in the world today (after rice, wheat, and maize), the bananas and plantains. These have always been a staple component of the human diet. Bananas and plantains are derived from the intra- and inter-specific hybridization of two wild diploid species Musa acuminata Colla (AA) and Musa balbisiana Colla (BB) (Simmonds and Shepherd Citation1955). However, Musa species are now grouped according to “ploidy,” the number of chromosome sets they contain, and the relative proportion of Musa acuminata (A) and Musa balbisiana (B) genes in their genome. Most familiar, seedless, cultivated varieties (cultivars) of banana are triploid hybrids (AAA, AAB, ABB). Diploids (AA, AB, BB) and tetraploids (AAAA, AAAB, AABB, ABBB) are much rarer; the latter essentially being experimental hybrids.
Banana is one of the oldest cultivated tropical fruits in India, where it grows from 0 to 1500 m asl. Among the main Indian states producing banana, West Bengal holds a key position, and the demand for banana cultivation and consumption has increased significantly in this state. Thus there is considerable need for improvement of molecular techniques for breeding which in turn requires a better understanding of the genome of the cultivated varieties or clones of edible Musa, thereby necessitating preparation of chromosomal database of the cultivars.
The genomes A from M. acuminata and B from M. balbisiana have been distinguished by the molecular genetic method of in situ hybridization (Osuji et al. Citation1997). Fluorescence in situ hybridization has also been used to localize ribosomal genes as well as telomere-like DNA sequences on the mitotic chromosomes of Musa species, cultivars and hybrids (Osuji Citation1998). Though cytological and cytogenetic work on Musa has reached an advanced stage, basic karyotype analysis is lacking in this genus. The unavailability of basic chromosome terminology or individual chromosomal characterization is lacking, owing to the small genome (Dolezel et al. Citation1994) and chromosome size (Osuji Citation1998; Osuji et al. Citation1996) in this genus. Another reason is the difficulty of obtaining good mitotic slides with plenty of metaphase cells (Osuji et al. Citation1996).
Fruits of cultivated Musa species are typically sterile or have extremely low fertility. They produce fruit pulp without pollination and fruits lacking seed (i.e. they are parthenocarpic). Although sterility and parthenocarpy are important factors that contribute to the desirability of banana fruits, sterility has impeded progress in breeding programs. Through natural somatic (vegetative) mutation, hybridization, and selection over many thousands of years, considerable genetic variability has arisen within the cultivated bananas, giving rise to more than 1000 varieties worldwide.
Systematic karyotype analysis of Musa cultivars of India is lacking. The aim of this work is to contribute to a karyomorphological repository for the cultivated accessions of edible Musa in West Bengal.
Materials and methods
Eight cultivars of edible Musa of West Bengal were collected from AICRP, BCKV, Mohanpur and maintained in the Experimental Garden of the Department of Botany, Presidency College, Kolkata. Young root tips were pretreated in 2 mM 8-hydroxyquinoline solution at RT for 2–2.30 hours, fixed in 3:1 acetoethanol overnight at 10–12°C, kept in 45% acetic acid for 25 minutes, hydrolyzed in 1 N HCl for 5 minutes at RT and stained with 2% acetoorcein. Fifteen metaphase plates per individual and five plants per cultivar were thoroughly examined under the microscope. The plates showing well-scattered metaphase were documented with the help of a ProgRes C3 (Jenoptik, Germany) camera attached to a Zeiss microscope (Jena, Germany). Lengths of the short and long arms of each chromosome were calculated. These values were then used to calculate the total chromatin length (TCL) and centromeric index (CI) of each cultivar. Karyotypes were constructed by arranging the chromosomes into groups on the basis of their centromeic index, ordering them by decreasing length within each category. The disparity index (DI) of chromosomes in a karyotype was calculated after Mohanty et al. (Citation1991) by the formula: DI = (longest chromosome – shortest chromosome/longest chromosome + shortest chromosome) × 100. The total form percentage or the mean centromeric index value (TF%) was calculated for each cultivar after Huziwara (Citation1962), by the following formula: TF% = (total sum of short arm length/total sum of chromosome length) × 100. Detection of the chromosomal differences was better using the karyotype arrangement.
Results
In the present study, we examined the eight cultivars of edible Musa listed in Table . Chromosome numbers of seven cultivars were found to be 2n = 3x = 33 and that of one cultivar (seeded banana) was found to be 2n = 2x = 22. The chromosomes were small (Osuji Citation1998), with a range of 1.00–2.97 μm in the cultivars studied. The disparity between the smallest and largest chromosomes was quite considerable. The chromosomes of the Musa cultivars reported in this study could be grouped into following three types:
Type A: Relatively long chromosomes with primary and secondary constrictions.
Type B: Relatively long to short chromosomes with median to nearly median primary constriction.
Type C: Relatively long to short chromosomes with sub median to nearly sub median primary constriction.
Table 1. Summary of cytogenetic results.
Descriptions of the metaphase chromosomes of individual cultivars are given below.
Cultivar 1: Amrit Sagar (AAA)
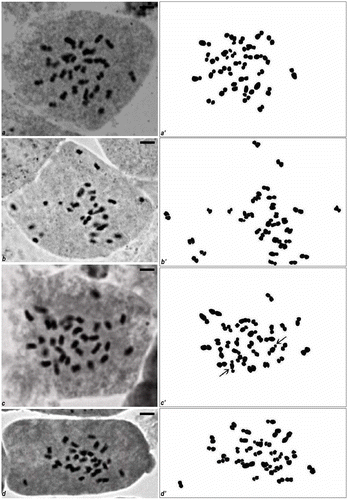
Accession no. BCKVV23. The chromosome number of Amrit Sagar was 2n = 3x = 33 (Figure , a′) with the karyotypic formula of 21B + 12C (Table ). Satellites were not found in the chromosomes of this cultivar in our study. The chromosome lengths were 1.3–2.48 μm. Of 21 chromosomes showing median to nearly median primary constriction (length 1.7–2.3 μm), 11 chromosomes were longer and the remaining 10 chromosomes were shorter in size. Submedian to nearly submedian primary constrictions were noted in 12 chromosomes but the length of this group varied from 1.3 to 2.5 μm; here two pairs of chromosomes were longer than the remaining four pairs (Figure ). The mean chromosome length of this cultivar was calculated to be 2.02 μm and TCL was 66.7.
Figure 2. Karyotypes of the eight cultivars of Musa: (a) Amrit Sagar; (b) Grand Naine; (c) Martaman; (d) Champa; (e) Kanthali Bagda; (f) Baish Chhara DT; (g) Baish Chhara CT; (h) seeded banana.

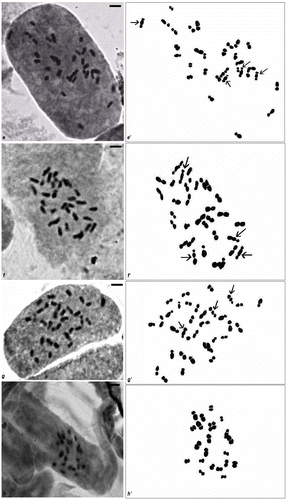
Figure 1. (a–d) Mitotic metaphase chromosomes in Musa cultivars; (a′–d′) hand drawings of mitotic metaphase plates of different cultivars. (e–h) Mitotic metaphase chromosomes in Musa cultivars; (e′–h′) hand drawings of mitotic metaphase plates of different cultivars.
Cultivar 2: Grand Naine (AAA)
Accession no. BCKVV23. The chromosome number of Grand Naine was 2n = 3x = 33 (Figure , b′) with the karyotypic formula of 24B + 9C (Table ). Satellites were not observed in the chromosomes of this cultivar. The chromosome lengths were 1.03–2.2 μm. Of 24 chromosomes showing median to nearly median centromere (length 1.04–2.2 μm), one pair of chromosomes was much smaller than the rest. Lengths of chromosomes with submedian to nearly submedian region centromere were 1.3–2.1 μm (Figure ). Mean chromosome length of the cultivar was 1.5 μm and TCL was 50.21.
Cultivar 3: Martaman (AAB)
Accession no. BCKVV32. The chromosome number of Martaman was 2n = 3x = 33 (Figure , c′) with the karyotypic formula of 2A + 22B + 9C (Table ). A pair of Type A chromosomes with satellite was noted in this cultivar. The chromosome lengths were 1.3–2.6 μm. The length of chromosomes with secondary constriction was 2.6 μm. The lengths of chromosomes with median to median region centromere were 1.32–2.17 μm; among the 22 chromosomes of this type two were much longer than the rest. The lengths of chromosomes with submedian to submedian region centromere were 1.7–2.14 μm; three chromosomes were much longer than the other six (Figure ). TCL was 58.7 and the mean chromosome length of this cultivar was calculated to be 1.8 μm.
Cultivar 4: Champa (AAB)
Accession no. BCKVV38. The chromosome number of Champa was 2n = 3x = 33 (Figure , d′) with the karyotypic formula of 22B + 11C (Table ). Satellites were not noted in this cultivar. The chromosome lengths were 1.32–2.55 μm. Most of the chromosomes were with median to median region centromere (22B) whose length were 1.32–2.55 μm; among these, four chromosomes were much longer, six chromosomes were much shorter and the length of the rest was intermediate (Figure ). Eleven chromosomes were with submedian to submedian region centromere; lengths were 1.6–2.14 μm. TCL was 57.3 and mean chromosome length was 1.74 μm.
Cultivar 5: Kanthali Bagda (ABB)
Accession no. BCKVV70. The chromosome number of Kanthali Bagda was 2n = 3x = 33 (Figure , e′) with the karyotypic formula of 4A + 23B + 6C (Figure ). Two pairs of Type A chromosomes with satellite were noted in this cultivar. The chromosome lengths were 1.2–2.3 μm. Of 23 chromosomes showing median to median region centromere lengths of 1.25–2.3 μm, two pairs of chromosomes were much longer than the rest. Lengths of chromosomes with submedian to submedian region centromere were 1.8–2.6 μm. The lengths of chromosomes with secondary constrictions were 2.1–2.6 μm. The mean chromosome length was 1.9 μm and TCL was calculated to be 63.7 (Table ).
Cultivar 6: Baish Chhara DT (ABB)
Accession no. BCKVV61. The chromosome number of Baish Chhara DT was 2n = 3x = 33 (Figure , f′) with the karyotypic formula of 4A + 20B + 9C (Figure ). Two pairs of Type A chromosomes with satellite were noted in this cultivar. The chromosome lengths were 1.5–2.5 μm. The lengths of chromosomes with median to median region centromere were 1.86–2.47 μm, and those of chromosomes with submedian to submedian region centromere were 1.45–2.08 μm. TCL was found to be 66.5 and mean chromosome length was 2.02 μm (Table ).
Cultivar 7: Baish Chhara CT (ABB)
Accession no. BCKVV74. The chromosome number of Baish Chhara CT was 2n = 3x = 33 (Figure , g′) with the karyotypic formula of 4A + 23B + 6C (Figure ). Two pairs of Type A chromosomes with satellite were noted in this cultivar. The chromosome lengths of this cultivar were 1.44–2.6 μm; the lengths of chromosomes with secondary constriction being 2.83 and 2.97 μm. The lengths of chromosomes with median to median region centromere were 1.44–2.6 μm and those of chromosomes with submedian to submedian region centromere were 1.6–2.09 μm. TCL was found to be 68.6 and mean chromosome length was 2.08 μm (Table ).
Cultivar 8: Seeded banana (BB)
Accession no. BCKVV10. The chromosome number of seeded banana was 2n = 2x = 22 (Figure , h′) with the karyotypic formula of 18B + 4C (Table ). Satellites were not noted in this cultivar. The chromosome lengths were 0.99–1.9 μm. The majority of the chromosomes (18B) showed median centromeres and their lengths were 0.99–1.9 μm; in this set two pairs of chromosomes were much longer than the rest (Figure ). Two pairs of chromosomes had centromere at the submedian position, with lengths 1.6–1.8 μm. The mean chromosome length of this cultivar was calculated to be 1.4 μm and TCL was 30.82.
Discussion
Musa spp. are an important crop in the subtropics and tropics and are derived from the intra- and inter-specific hybridization of two wild diploid species, Musa acuminata Colla (AA) and Musa balbisiana Colla (BB) (Simmonds and Shepherd Citation1955). Inter-crossing among species and subspecies has resulted in the appearance of sterility, a trait that was selected during domestication, together with parthenocarpy and vegetative propagation (Simmonds Citation1995). The first cultivated bananas were diploids, and triploid forms originated from the crossing between partially sterile diploids with fertile male forms. Since triploids were more productive and vigorous, with larger fruits, they were selected over the diploid clones (Sharrock Citation1998). Conventional methods of breeding are complicated due to the triploid nature and only a few diploid clones produce viable pollen. Improvement for disease resistance and productivity requires the use of biotechnological tools. Consequently, plantains and bananas (Musa) have become the subjects of intense improvement programs in which modern biotechnological methods have played significant roles. Some of these techniques, however, have been reported to predispose plant materials to chromosomal instability. This does not preclude the genomic instability that ordinarily arises due to cryptic chromosomal rearrangements, somatic crossing over with sister chromatid exchanges, transposable elements, and gene amplification/diminution phenomena (Mantell et al. Citation1985; Hartwell et al. Citation2000). The celibacy of the cultivated bananas makes them scientifically interesting, as there is no genetic exchange during reproduction, but random mutations occur and may be selected. In order to understand banana genetics more, knowledge must be gained about the karyomorphological characters of the edible cultivars.
Edible bananas have 22, 33 or 44 chromosomes, representing diploid, triploid and tetraploid cultivars (Stover and Simmonds Citation1987). These cultivars have a wide range of genome permutations, including AA, AB, BB, AAA, AAB, ABB, AAAB, ABBB, and AABB. Based on morphological observations of the characters that differentiate the two wild species and on the ploidy level of the different clones, Simmonds and Shepherd (Citation1955) recognized five main genomic groups of cultivated bananas, designated AA, AB, AAA, AAB, and ABB. Within each group, related clones are associated in a subgroup. Studies have been carried out on Musa chromosomes but these are restricted to African locations. Chromosomal studies on edible Musa of India have not been attempted to date. In our study, we have started extensive studies on Musa chromosomes of the edible varieties of West Bengal with a desire to extend the study to the cultivars of India in the near future. The small size of the genome (Dolezel et al. Citation1994) and chromosomes (Isobe and Hashimoto Citation1994; Osuji et al. Citation1996) and the difficulty in obtaining dividing cells (Osuji et al. Citation1996) have made cytological study difficult.
The fact that the basic chromosome number of the section Musa is x = 11 has been reconfirmed in the present study. We had prior information about the genome composition of the collected cultivars from our collection source (AICRP, BCKV, Mohanpur). The seedless cultivars were found to be triploid when chromosomes were counted and the seeded banana was diploid.
The karyotypes reported in this work consist mainly of chromosomes with median, nearly median, submedian or nearly submedian position of centromere. Significant variations in chromosome were not noted while analyzing the karyotypes of the eight cultivars studied. The seeded banana, which is supposed to represent one of the wild genomes of Musa from which the different varieties may have originated, consists of chromosomes with median and submedian constrictions. The remaining seven triploid varieties known to have been derived from hybridization of the wild species have almost similar combinations of chromosomes with median and submedian constrictions, with minute variations. Additional types of chromosomes (with median region and submedian region centromere) have been found during our study in these cultivars. Studies of the karyomorphology of bananas and plantain, which have many cultivars and subgroups, are lacking mainly because of the small size of the chromosomes and the difficulty in obtaining a sufficient number of cells containing metaphase chromosomes, as also reported by earlier researchers. In the present study we have performed several trials with the methods of pretreatment and fixation and hydrolysis (Osuji Citation1998; Osuji et al. Citation2006) before reporting the final method.
Cytological data are essential to the study of plant evolution and diversification (Stebbins Citation1950, Citation1971; Hong Citation1990; Stace Citation2000). Chromosome size and morphology may help indicate evolutionary relationships among plant species (Clark and Wall Citation1996). Despite stability in chromosome number, large variations in chromosome size have been found to play an important role in the evolution of Lathyrus species (Seijo and Fernández Citation2003). Differences in karyotype formulae among the different cultivars of edible Musa suggest that structural changes may have contributed to the diversification of the bananas. However, the fact that variations are minor indicates that if the mechanisms of speciation did involve chromosome rearrangements, these may not have been large structural mutations, but small or cryptic changes.
Consequently, in a detailed analysis of karyotypes, we did not find any chromosome features that were diagnostic for the cultivars, but a combination of features – chromosome mean length, presence of SAT chromosomes, type of SAT chromosomes, and karyotype formula – allows us to differentiate the varieties. The standardization of karyotype analysis would open new vistas for characterization of the chromosomes of different cultivars and subgroups through banding and more advanced cytogenetic techniques.
Acknowledgment
The authors are grateful to University Grants Commission, India for providing financial aid, BCKV, India for providing plant material and the Department of Botany, Presidency University, Kolkata, India for providing laboratory facilities.
References
- Clark MS, Wall WJ. 1996. Chromosomes, the complex code. London: Chapman & Hall, p. 237–243.
- Dolezel J, Dolezelova M, Novak FJ. 1994. Flow cytometric estimation of nuclear DNA amounts in diploid bananas (Musa acuminata and M. balbisiana). Biol Plantarum. 36: 351–357.
- Hartwell LH, Hood L, Goldberg ML, Reynolds AE, Silver LM, Veres RC. 2000. Genetics: From gene to genomes, the chromosome theory of inheritance. High Educ. 3:70–90.
- Hong DY. 1990. Plant cytotaxonomy. Beijing: Science Press.
- Huziwara Y. 1962. The karyotype analysis in some genera of Compositae X. The chromosomes of some European species of Aster. Bot Mag. 75:143–150.
- Isobe M, Hashimoto K. 1994. The chromosome count of nine taxa in Musa and its allied genus Musella. Bull Hiroshima Bot Gard. 15:7–11.
- Mantell SH, Matthews JA, McKee RA. 1985. Principles of plant biotechnology. An introduction to genetic engineering in plants. London: Blackwell Scientific Publications.
- Mohanty BD, Ghosh PD, Maity S. 1991. Chromosomal analysis in cultured cells of barley (Hordeum vulgare L.) Structural alterations in chromosomes. Cytologia. 56:191–197.
- Osuji JO. 1998. Chromosomal investigations of infra-specific taxa in Musa paradisiaca L. (Plantain; Musa spp. AAB group). J Innov Life Sci. 3:87–92.
- Osuji JO, Crouch J, Harrison G, Heslop-Harrison JS. 1997. Identification of the genomic constitution of Musa L. lines (bananas, plantains and hybrids) using molecular cytogenetics. Ann Bot. 80:787–793.
- Osuji JO, Crouch J, Harrison G, Heslop-Harrison JS. 1998. Molecular cytogenetics of Musa species, cultivars and hybrids: location of 18S-5.8S-25S and 5S rDNA and telomere-like sequences. Ann Bot. 82:243–248.
- Osuji JO, Okoli BE, Ortiz R. 1996. An improved procedure for mitotic studies of the Eumusa section of the genus Musa L. (Musaceae). Infomusa. 5:12–14.
- Osuji JO, Okuli BE, Hilary OE. 2006. Karyotypes of the A and B genomes of Musa L. Cytologia. 71 (1): 21–24.
- Seijo JG, Fernádez A. 2003. Karyotype analysis and chromosome evolution in South American spcies of Lathyrus (Leguminosae). Am J Bot. 90:980–987.
- Sharrock S. 1998. Collecting the Musa gene pool in Papua New Guinea. In: Guarino L, Rao VR, Reid R, eds. Collecting plant genetic diversity. Wallingford: CAB International, 647–658.
- Simmonds NW. 1995. Banana: Musa (Musaceae). In: Smartt J, Simmonds NW, eds. Evolution of crop plants. 2nd ed. Essex: Longman Scientific & Technical, p. 370–375.
- Simmonds NW, Shepherd K. 1955. The taxonomy and origins of the cultivated bananas. Bot J Linn Soc. 55:302–312.
- Stace CA. 2000. Cytology and cytogenetics as a fundamental taxonomic resource for the 20th and 21st centuries. Taxon. 49:451–477.
- Stebbins GL. 1950. Variation and evolution in plants. New York: Columbia University Press.
- Stebbins GL. 1971. Chromosomal evolution in higher plants. London: Edward Arnold.
- Stover RH, Simmonds NW. 1987. Bananas. 3rd ed. Essex: Longman Scientific and Technical, p. 468.