Abstract
Chromosome length is a basic karyotype characteristic. This paper shows, on the basis of total chromosome length (TCL) measurement, that this value is not a stable one. TCL varies among different c-metaphases from one meristem; the average TCL is different for (a) different pre-treatment agents; (b) different pre-treatment times for the same pre-treatment agents; and (c) between siblings treated simultaneously. In the same way, total chromosome volume (TCV) is not a suitable characteristic for chromosome size comparison. To solve the problem of chromosome size comparison the following standardization of TCL measurement is proposed: to determine TCL in c-metaphases with the chromosomes contracted to maximum level and still retaining all measurable karyotype characteristics. This way of standardization requires a pre-treatment method providing c-metaphases suitable for standardized TCL measurement. The paper also brings one new chromosome number to science: Onosma thracica, 2n = 14.
Introduction
Classical karyological methods – studies of chromosome characteristics using coloured squashes made of meristematic parts of plants – represent simple, quick methods usable by a wide range of scientists without need of special equipment. They provide important data on chromosome size, morphology and further karyotype characteristics. For these reasons they retain, even in this time of modern cytogenetic and molecular approaches, an important role in taxonomic and biosystematic studies and are still widely used (e.g. López et al. Citation2009; Klos et al. Citation2009; Peruzzi et al. Citation2009; Tanaka et al. Citation2009; Kazem et al. Citation2010; Ray Citation2010; Zhang, Zhang et al. Citation2010; Ghasem et al. Citation2011; Sheidai et al. Citation2011; Sosa et al. Citation2011; Zhang et al. Citation2011; Zhou et al. Citation2011; Ebrahim et al. Citation2012; Liu et al. Citation2012; Kiran et al. Citation2012; Vega and Dematteis Citation2012; Uslu 2013; Genç et al. Citation2013; Viciani et al. Citation2013; Chung et al. Citation2013).
Particular karyotype characteristics – especially chromosome number, chromosome length, total chromosome/complement length (TCL, the sum of lengths of all chromosomes of one nucleus), relative length of particular chromosomes (the ratio of individual chromosome length to TCL), arm ratio (the ratio of lengths of longer to shorter arm in particular chromosomes), presence, number and position of satellites – are often compared to find the relationship between different taxa or differences within a taxon or for other biosystematic reasons (e.g. Mikoláš and Mihoková Citation1995; Naranjo et al. Citation1998; Mártonfiová Citation2004; Mártonfi et al. Citation2008; Peruzzi et al. Citation2009; Rosato and Rosselló Citation2009; Tanaka et al. Citation2009; Conterato et al. Citation2010; Corrêa et al. Citation2010; Tarinejada and Mirshekari Citation2010; Zhang, Zhang et al. Citation2010; Arabbeigi et al. Citation2011; Ghaffaripour et al. Citation2011; Ghasem et al. Citation2011; Mehes-Smith et al. Citation2011; Sosa et al. Citation2011; Zhang et al. Citation2011; Anjali and Srivastava Citation2012; Cheema and Pant Citation2013; Aksu et al. Citation2013; Wang et al. Citation2013).
The lengths of particular chromosomes and their parts are usually measured in selected (“best”) metaphase plates (Tanaka et al. Citation2009; Hejazi Citation2011; Ebrahim et al. Citation2012) found in the slides prepared from plant meristems pre-treated by different mitotoxic substances (Das et al. Citation1998a; Jena et al. Citation2003; Mohanty and Das Citation2006; López et al. Citation2009; Rosato and Rosselló Citation2009; Conterato et al. Citation2010; Zhang, Wu et al. Citation2010; Zhang, Zhang et al. Citation2010; Hejazi Citation2011; Mehes-Smith et al. Citation2011; Zhang et al. Citation2011). These selected metaphase plates are characterized by well-spread chromosomes, in which all elements necessary for chromosome characterization are identifiable. Usually, the number of analysed c-metaphases from one sample is not high (usually 5–10); sometimes chromosome characteristics are calculated from a single selected c-metaphase plate (Dvořák Citation1979). However, particular metaphase plates may differ by a degree of contraction (condensation and spiralization) of chromosomes they contain.
Metaphase chromosome size can be expressed by different characteristics. Chromosome length and TCL are employed the most often. Another characteristic, chromosome volume, was introduced by De Vescovi and Sziklai (Citation1975) and approximates the chromosome by a cylinder with radius r and height h (Figure ). Chromosome volume is then calculated by v = πr2h. TCV (total chromosome volume) has been employed much less than TCL (White and Rees Citation1987; Sharma and Ramesh Citation1997; Das et al. Citation1998a; Naranjo et al. Citation1998; Jena et al. Citation2003; Mohanty and Das Citation2006; Poggio et al. Citation2007; Ghasem et al. Citation2011). Nuclear DNA content is another characteristic that is related to chromosome size.
Chromosome length and especially total chromosome length is a characteristic often used to compare two very similar taxa (e.g. Mekki et al. Citation2007; López et al. Citation2009; Mehes-Smith et al. Citation2011). Many studies ignore the fact that chromosome length is not stable (Jena et al. Citation2003; Peruzzi et al. Citation2009; Corrêa et al. Citation2010; Ghasem et al. Citation2011; Hejazi Citation2011). Das et al. (Citation1998a, Citation1998b) noted that differences in chromosome length among species may be due to different condensation and spiralization of chromosomes. A comparison of TCL without sufficient care can lead to incorrect interpretations. However, TCL is important karyotype characteristic that should not be ignored.
There are species or even genera or groups of genera with identical karyotypes, in which nuclear DNA amount varies substantially despite karyotypical uniformity (Brandham and Doherty Citation1998; Poggio et al. Citation2007). This problem also leads to the need for comparison of TCL in the taxa concerned.
This work is aimed at (1) showing different factors influencing TCL; (2) standardizing the measurement of TCL, so that the values are comparable for different taxa and different authors and different classical karyological methods; and (3) obtaining insights into the problem of the relationship between TCL and nuclear DNA content value.
Materials and methods
Karyology
In order to avoid an influence of genetic variability on chromosome size, for basic measurements the roots with root-tip meristems were taken all at once from a single onion plant (Allium cepa L. cv. Radar), in which the growth of adventive roots had been provoked (leaving it on beaker filled with water) several days prior to the sample taking. To compare the different methods, the roots were treated by one of the following pre-treatments:
0.01% aqueous solution of colchicine at room temperature for (a) 1 hour; (b) 2 hours; and (c) 3 hours.
0.002 M solution of 8-hydroxyquinoline at 2–4°C for 18 hours (overnight).
Saturated aqueous solution of p-dichlorobenzene at room temperature for (a) two hours; and (b) three hours.
Fixation was in a 3:1 mixture of 96% ethanol and glacial acetic acid, material was macerated by 1 N HCl at 60°C for 5 minutes, washed in distilled water, and meristems were then squashed by a cellophane square technique (Murín Citation1960). The slides were stained by a 7% solution of Giemsa stain, modified solution (Fluka Analytical, Switzerland) in distilled water, dried and observed in a drop of immersion oil. Each slide contained material only from one root. Suitable c-metaphase plates for karyotype analyses (i.e. metaphase plates with well-spread chromosomes in which primary and secondary constrictions were unambiguously detectable) were selected. Ten suitable c-metaphase plates (or less, if less then 10 were present in the slide) were selected randomly, as they were gradually found in the slide (except for one special c-metaphase plate intended for standardization of chromosome length measurement, which was selected for purpose). The selected c-metaphase plates were photographed (using a Leica DM 2500 microscope equipped with camera DFC 290 HD and software Leica application suite version 3.5.0, Switzerland), individual chromosomes were measured and chromosome length, or chromosome volume (De-Vescovi et Sziklai 1975) was calculated.
To compare different plant taxa from the point of view of the relationship between TCL and DNA content, the following plant material was employed: Picris hispidissima W.D.J. Koch, Taraxacum sect. Ruderalia Kirschner, H. Øllg. et Štěpánek, Onosma thracica Vel., Trifolium pratense L., Galium verum L., Leontodon hispidus L., Tragopogon orientalis L., Pimpinella saxifraga L., Lycopersicon esculentum Mill., and Vicia faba L. (detailed data on these plants are given in Table ). In Picris hispidissima siblings were also compared from the point of view of TCL variation. Root tip meristems of these plants (either potted plants or seedlings) were pre-treated with a 0.002 M solution of 8-hydroxyquinoline overnight at 2–4°C and the slides were prepared according to the same method as described above.
Table 1. Species included in this study with chromosome numbers, data on collection in the case of wild species, cultivar data in the case of cultivated species.
Flow cytometry
The samples for flow cytometry analysis were prepared from the leaves of the plants by a two-step procedure, consisting of separate nuclear isolation and staining steps, using propidium iodide as DNA intercalator (Otto 1990; Doležel and Göhde Citation1995). DNA contents were measured on Partec CyFlow ML (Partec, Münster, Germany) flow cytometer equipped with green laser (532 nm/150 mV) in the Laboratory of Flow Cytometry at the Institute of Biological and Ecological Sciences of P. J. Šafárik University in Košice (Slovakia). As an internal standard, Pisum sativum Ctirad was used in the case of Allium cepa, Tragopogon orientalis and Pimpinella saxifraga; Lycopersicon esculentum “Stupické tyčkové polní” in the case of Taraxacum and Picris hispidissima; Glycine max “Polanka” in the case of Galium verum; Zea mays CE-777 in the case of Leontodon hispidus. Approximately 1 cm2 of young leaf of sample and standard was chopped with a new razor blade (in a Petri dish) in 1 ml of cold Otto I buffer (100 mM citric acid, 0.5 % (v/v) Tween 20 (pH 2–3)) and this suspension was filtered through 42 μm nylon mesh and centrifuged. Supernatant was removed and pellet (c.100 μl) was resuspended in 100 μl of fresh Otto I buffer. After 30 min of incubation at room temperature, 1ml of fluorochrome solution was added. It consisted of Otto II buffer (400 mM Na2PO4·12H2O (pH 8–9)), and propidium iodide (50 μg per sample), RNAse (50 μg per sample) and β-mercaptoetanol (2 μl per sample). After 10 min incubation at 4°C the intensity of fluorescence of the samples was measured at the flow cytometer. For each sample 5000 nuclei were analysed using FloMax software (version 2.70).
Results and discussion
The chromosome number of the material employed in the study is given in the Table . In order to avoid mistakes connected with identification of particular chromosomes, total chromosome length (TCL) measurement was employed to express variation of chromosome length. TCL varies between different “best” c-metaphases within one meristem (obtained from the same root), see Tables , and Figure .
Table 3. TCL values for different c-metaphases coming from one meristem of Allium cepa (pre-treated with 0.01% colchicine for 3 h), one meristem of Taraxacum linearisquameum and one meristem of Picris hispidissima (pre-treated by 0.002 M solution of 8-hydroxyquinoline overnight at 2–4°C).
Table 2. Survey of TCL measurements for different karyological pre-treatment methods employed in Allium cepa.
Figure 1. Approximation of chromosome by a cylinder: r, radius of the cylinder, represents half the measured chromosome width; h, chromosome length.
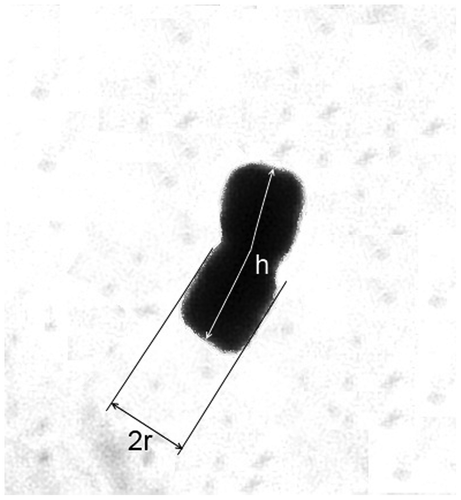
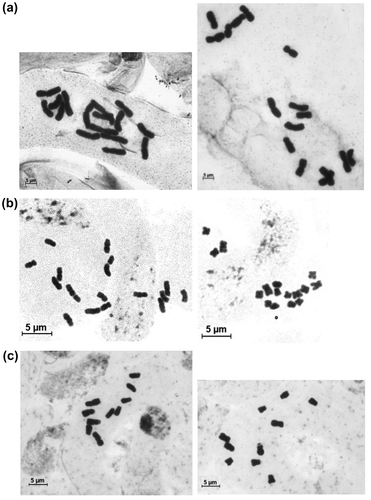
Figure 2. C-metaphases with different TCL coming from the same meristem of (a) Allium cepa L. (TCL 169.12 μm and 133.96 μm); (b) Taraxacum linearisquameum (TCL 43.2 μm and 25.57 μm); (c) Picris hispidissima (TCL 30.49 μm and 24.86 μm).
Average TCL (calculated as an arithmetical average of TCL for the selected c-metaphases) is different for different pre-treatment agents; see Table . The time of exposition of a meristem to certain pre-treatment agent influences the chromosome length as well – prolongation of the time of the exposure causes further shortening of the chromosomes; see Table , Figure . However, exceeding a certain limit of exposure time causes damage of chromosomes and c-metaphases (Figure ).
Figure 4. Damage of c-metaphases of Allium cepa caused by a too-long exposure to pre-treatment (saturated solution of p-dichlorobenzene for 18 h).]
![Figure 4. Damage of c-metaphases of Allium cepa caused by a too-long exposure to pre-treatment (saturated solution of p-dichlorobenzene for 18 h).]](/cms/asset/d44e6af0-2fe3-49b7-8fec-4f3b27d0f41c/tcar_a_854565_f0004_b.gif)
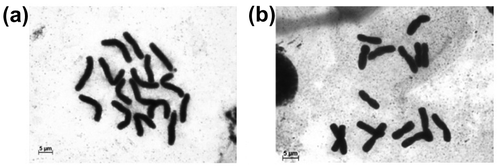
Figure 3. C-metaphases of Allium cepa pre-treated with 0.01% colchicine solution (a) for 1 hour; (b) for 3 hours. C-metaphases with the closest TCL value to the average one for particular pre-treatment times were selected.
Chromosome length differs also between siblings – average TCL for seedlings coming from the same capitulum of Picris hispidissima, which were simultaneously karyologically treated, was 28.18 μm (SD 2.03) and 32.57 μm (SD 2.17).
Chromosome length can be influenced by many other (often poorly recognized) factors connected with both plant material studied and methodology employed. For example, younger meristems can react otherwise to the same method than the older ones, and some karyologists prepare their slides by accessive pressing of macerated meristems, which would influence not only the total chromosome length, but can cause unequal length changes among and also within particular chromosomes and thus shift calculated karyotype characteristics.
It is clear from the above measurements that TCL is not a suitable variable for comparison of chromosome size and making conclusions about different taxa, populations, etc., as it is often used (Das et al. Citation1998a, Das et al. Citation1998b; Mekki et al. Citation2007; Corrêa et al. Citation2010; Hejazi Citation2011). However, such comparison is often desired.
TCV (total chromosome volume) is another variable characterizing chromosome size. TCV (the sum of volumes of individual chromosomes of the complement) varied between different c-metaphases within one slide and average TCV was different for different methods employed (Table ). In order to reach better accuracy, in the case of Picris hispidissima longer and shorter arms of individual chromosomes were approximated by a separate cylinder. TCV cannot be recommended as a suitable variable for chromosome size comparison. Modified TCV, which would approximate chromatids by two separate cylinders would lead to a value for the volume of particular chromosomes of v(m) = 2π(r/2)2h = v/2, where v is the volume of chromosome approximated by one cylinder; and thus to the same variation. Comparison of chromosome size by means of TCL differs from that done by means of TCV; higher TCV does not imply higher TCL and vice versa (see Table ). This has also been confirmed by other researchers (Mohanty and Das Citation2006; Das Citation2008). TCV is also not consistent with nuclear DNA content (Brandham and Doherty Citation1998; Mohanty and Das Citation2006).
Table 4. TCV and TCL values for individual c-metaphases and average TCV values for (a) two different pre-treatment methods in Allium cepa; (b) Picris hispidissima.
Chromosome size measurement standardization seems not to have a simple solution. The standardization may issue from the fact that metaphase chromosomes reach a certain maximum level of contraction during mitosis. Therefore, measurement of TCL in selected c-metaphases where chromosomes are contracted to a maximum degree is a possible way to measure “standardized” TCL. For this measurement, a suitable pre-treatment method that provides sufficiently contracted chromosomes must be found. The chromosomes should be contracted as much as possible without any damage by a too-strong pre-treatment (too-long pre-treatment time or too-high concentration of pre-treatment agent) and should retain all the measurable karyotype characteristics. In this way, the metaphases selected for standardized TCL measurement represent only a subset of all c-metaphases in which karyotype characteristics are measured, or additional suitable c-metaphases should be chosen specially for standardized TCL measurement.
For Allium cepa all of the three commonly used karyological pre-treatment methods, with suitable pre-treatment time, provided c-metaphases with sufficiently contracted chromosomes suitable for standardized TCL value measurement (Table ).
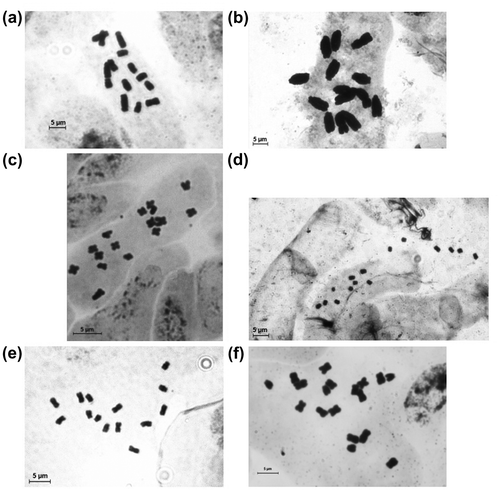
An attempt to measure standardized TCL for both Picris hispidissima and Taraxacum linearisquameum provided satisfactory results. The employed method provided, besides the others, the c-metaphases suitable for standardized TCL measurement – these are the ones with the most contracted chromosomes showing nearly the same TCL value (see Table ; the first three values for T. linearisquameum and first two values for P. hispidissima). An example of c-metaphases suitable for standardized TCL measurement for different taxa is presented in Figure .
Figure 5. An example of c-metaphases suitable for standardized TCL measurement for different taxa: (a) Leontodon hispidus; (b) Vicia faba; (c) Taraxacum linearisqumeum; (d) Trifolium pratense; (e) Onosma thracica; (f) Matricaria chamomilla cv. “Novbona”.
Karyotype studies of species with the same chromosome number and very similar karyotype characteristics but with different genome sizes, which require TCL standardization, will be presented in another paper as part of a wider taxonomical study. However, a similar problem was met by Brandham and Doherty (Citation1998) who solved the mechanism of the evolutionary increase of DNA amount when the karyotype morphology remains stable. They suggest that evolutionary increase in DNA amount involves the amplification of many small DNA segments at sites distributed evenly throughout the chromosome complement, in numbers proportional to chromosome length, thus maintaining karyotypic uniformity in the genus.
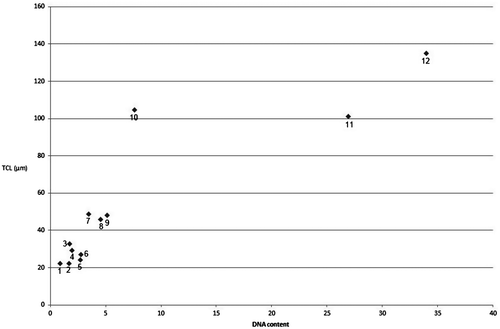
The relationship between standardized TCL and nuclear DNA content in several different species is given in Table and Figure . This relationship is not linear; however, there is some suggestion that, with some exceptions, an increase in DNA content implies an increase in TCL. Papers presenting both TCL and genome size or nuclear DNA content are rare. Peruzzi et al. (Citation2009) gives both average THL (total haploid length, half of TCL value) and average genome size for numerous genera within the family Liliaceae. The relationship between them is similar to those in our work, presented in Figure . Mekki et al. (Citation2007) find an almost linear relationship between the total chromosome length and nuclear DNA content for different cultivars of Phaseolus vulgaris. Lee et al. (Citation2004) study the possibility of approximation of nuclear 2C DNA content by certain chromosome characteristics in some wheat and rye species, and show correlation between nuclear 2C DNA content and total chromosome length. However, these data should be interpreted with care since different degrees of chromosome contraction were not taken into consideration in these papers, similarly to other papers with such data (Das et al. Citation1998a, Citation1998b). However, it is quite probable that for very close taxa the relationship between DNA content and standardized TCL approximates a linear one. This problem requires further studies.
Table 5. Standardized TCL and 2C DNA content for different species.
Figure 6. A relationship between 2C DNA content and standardized TCL for several species: 1. Trifolium pratense; 2. Taraxacum linearisquameum; 3. Galium verum; 4. Lycopersicon esculentum “Stupické”; 5. Picris hispidissima; 6. Onosma thracica; 7. Taraxacum sect. Ruderalia – tetraploid; 8. Leontodon hispidus; 9. Tragopogon orientalis; 10. Pimpinella saxifraga; 11. Vicia faba “Inovec”; 12. Allium cepa.
Conclusions
TCL measurement standardization is necessary for any study comparing chromosome size of different taxa. The proposed standardization – determination of TCL in c-metaphases with the chromosomes contracted to maximum level and still retaining all measurable karyotype characteristics – represents possible solution to this problem. However, selection of suitable karyological methodology and suitable c-metaphases for this measurement requires certain experience with karyological methods and chromosome measurement and thus results achieved by different scientists should be compared with care.
Acknowledgements
Support for this research was provided by APVV Grant Agency (APVV 0320-10), and VEGA grant agency (VEGA 1/0173/11). My thanks are also due to Vladislav Kolarčíik and Valeria Kocovaá (Faculty of Science, P.J. Safarik University, Košice) for providing material and data on genome size of Onosma thracica and Trifolium pratense, respectively, to Marek Slovák (Botanical Institute of Slovak Acad. Sci., Bratislava) for providing material of Picris hispidissima and to Pavol Mártonfi (Faculty of Science, P.J. Safarik University, Košice) for identification of several taxa.
References
- Aksu N, Inceer H, Hayirlioǧlu-Ayaz S. 2013. Karyotype analysis of six Achillea L. (Asteraceae, Anthemideae) taxa from Turkey. Caryologia. 66(2): 103–108.
- Anjali M, Srivastava AK. 2012. Karyological studies in twelve accessions of Carthamus tinctoriusi. Caryologia. 65(1): 1–6.
- Arabbeigi M, Arzani A, Saeidi G. 2011. Study of karyotype and nucleolar organizer regions (NORs) in wild, synthetic and cultivated wheats. Emirates J Food Agric. 23(2): 196–203.
- Brandham PE, Doherty M-J. 1998. Genome size variation in the Aloaceae, an angiosperm family displaying karyotypic orthoselection. Ann Bot. 82(Supplement A): 67–73.
- Cheema SK, Pant MR. 2013. Karyotype analysis of seven cultivated varieties of Capsicum annuum L. Caryologia. 66(1): 70–75.
- Chung K-S, Oh B-U, Park MS, Nam BM, Chung GY. 2013. Chromosome numbers of 28 taxa in 10 genera of the Ranunculaceae (buttercup family) from the Korean peninsula. Caryologia. 66(2): 128–137.
- Conterato IF, Schifino-Wittmann MT, Dall’Agnol M. 2010. Seed dimorphism, chromosome number and karyotype of the amphicarpic species Trifolium argentinense Speg. Genet Resour Crop Evol. 57(5): 727–731.
- Corrêa AM, Jung-Mendaçolli SL, Forni-Martins ER. 2010. Karyotype characterisation of Brazilian species of the genus Psychotria L. – subfamily Rubioideae (Rubiaceae). Kew Bull. 65(1): 45–52.
- Das AB. 2008. Assessment of genetic diversity and phylogenetic analysis of ‘star cactus’ (Astrophytum) through chromosomes and RAPD markers. Cytologia. 73(2): 179–188.
- Das AB, Mohanty S, Das P. 1998a. New report on chromosome number, karyotype and 4C DNA content in three species of Pachypodium Lindley. Caryologia. 51(3–4): 245–252.
- Das AB, Mohanty S, Das P. 1998b. Interspecific variation of nuclear DNA and structural changes in meiotic and mitotic chromosome in some species of Mammillaria (Cactaceae). Caryologia. 51(3–4): 289–301.
- De Vescovi MA, Sziklai O. 1975. Comparative analysis of Douglas fir, Pseudotsuga menziesii (Mirb.) Franco. Silvae Genetica. 24(2–3): 68–73.
- Doležel J, Göhde W. 1995. Sex determination in dioecious plants Melandrium album and M. rubrum using high-resolution flow cytometry. Cytometry. 19: 103–106.
- Dvořák F. 1979. Study of chromosomes of Angiosperms 8. Scripta Facultatis Scientiarium Naturalium. UJEP Brunensis, Biology. 9(2): 85–103.
- Ebrahim F, Pakniyat H, Arzani A, Rahimmalek M. 2012. Karyotype analysis and new chromosome number reports in Achillea species. Biologia. 67(2): 284–288.
- Genç I, Özhatay N, Cevri M. 2013. A karyomorphological study of the genus Allium (sect. Melanocrommyum) from Turkey. Caryologia. 66(1): 31–40.
- Ghaffaripour S, Karimizadeh R, Mohammadi M, Suq HS, Shefazadeh MK, Mohammadi N. 2011. Karyotypic studies on some genotypes of hull-less barley (Hordeum vulgare L.). Turk J Field Crop. 16(2): 245–250.
- Ghasem K, Danesh-Gilevaei M, Aghaalikhani M. 2011. Karyotypic and nuclear DNA variations in Lathyrus sativus (Fabaceae). Caryologia. 64(1): 42–54.
- Hejazi SMH. 2011. Karyological study on three Cicer L. species (Fabaceae) in Iran. Asian. J Cell Biol. 6(3): 97–104.
- Jena S, Sahoo P, Das AB. 2003. New reports of chromosome number and genome size in eight mangroves from coastal Orissa. Caryologia. 56(3): 353–358.
- Kazem Y, Houshmand S, Dadane GZ. 2010. Karyotype analysis of Astragalus effusus Bunge (Fabaceae). Caryologia. 63(3): 257–261.
- Kiran Y, Turkoglu I, Kirilmaz F, Arabaci T, Sahin A, Bagci E. 2012. Karyological investigation of six Achillea L. (Asteraceae) species growing in Turkey. Caryologia. 65(2): 101–105.
- Klos J, Sliwinska E, Kula A, Golczyk H, Grabowska-Joachimiak A, Ilnicki T, Szostek K, Stewart A, Joachimiak AJ. 2009. Karyotype and nuclear DNA content of hexa-, octo-, and duodecaploid lines of Bromus subgen. Ceratochloa. Genet Mol Biol. 32(3): 528–537.
- Lee J, Ma Y, Wako T, Li LCh, Kim K, Park S, Uchiyama S, Fukui K. 2004. Flow karyotypes and chromosomal DNA contents of genus Triticum species and rye (Secale cereale). Chromosome Res. 12(1): 93–102.
- Liu Y, Deng T, Yang Q-E. 2012. Karyology of the genus Faberia (Cichorieae-Asteraceae) and its systematic implications. Nordic J Bot. 30 (3): 365–371.
- López MG, Wulff AF, Poggio L, Xifreda CC. 2009. South African fireweed Senecio madagascariensis (Asteraceae) in Argentina: relevance of chromosome studies to its systematics. Bot J Linn Soc. 158(4): 613–620.
- Mártonfi P, Mártonfiová L, Kolarčik V. 2008. Karyotypes and genome size of Onosma species from northern limits of the genus in Carpathians. Caryologia. 61(4): 363–374.
- Mártonfiová L. 2004. Karyotype studies in Pulsatilla zimmermannii. Biologia. 59(1): 61–64.
- Mehes-Smith M, Nkongolo KK, Kim NS. 2011. A comparative cytogenetic analysis of five pine species from North America, Pinus banksiana, P. contorta, P. monticola, P. resinosa, and P. strobus. Plant Syst Evol. 292(3–4): 153–164.
- Mekki L, Badr A, Fekry M. 2007. Cytogenetic studies on nine genotypes of Phaseolus vulgaris L. cultivated in Egypt in relation to zinc efficiency. Pakistan. J Biol Sci. 10(23): 4230–4235.
- Mikoláš V, Mihoková L. 1995. Diploidi a polyploidi sekce Ruderalia Kirschner, Olgaard et Štěpánek rodu Taraxacum na Slovensku: Příspěvek k jejich studiu. Zborník z 5. zjazdu SBS, Blatnica, 92–97.
- Mohanty S, Das AB. 2006. Interspecific genetic diversity in 15 species of Cassia L. evident by chromosome and 4C nuclear DNA analysis. J Biol Sci. 6(4): 664–670.
- Murín A. 1960. Substitution of cellophane for glass covers to facilitate preparation of permanent squashes and smears. Stain Technol. 35: 351–353.
- Naranjo CA, Ferrari MR, Palermo AM, Poggio L. 1998. Karyotype, DNA content and meiotic behaviour in five South American species of Vicia (Fabaceae). Ann Bot. 82(6): 757–764.
- Otto FJ. 1990. DAPI staining of fixed cells for high-resolution flow cytometry of nuclear DNA. In: Darzynkiewicz Z, Crissman HA, eds. Methods in cell biology. San Diego: Academic Press, 33: 105–110.
- Peruzzi L, Leitch IJ, Caparelli KF. 2009. Chromosome diversity and evolution in Liliaceae. Ann Bot. 103(3): 459–475.
- Poggio L, González G, Naranjo CA. 2007. Chromosome studies in Hippeastrum (Amaryllidaceae): variation in genome size. Bot J Linn Soc. 155(2): 171–178.
- Ray DP. 2010. Karyotype and genome size variation in five new cultivars of Indian Brinjal (Solarium melongena var. Esculentum L.). Cytologia. 75(1): 41–47.
- Rosato M, Rosselló JA. 2009. Karyological observations in Medicago section Dendrotelis (Fabaceae). Folia Geobot. 44(4): 423–433.
- Sharma NK, Ramesh B. 1997. Karyotype analysis in faba bean. Ann Arid Zone. 36(4): 363–366.
- Sheidai M, Eftekharian R, Gholipoor A, Noormohammadi Z. 2011. Population diversity and polyploidy incidence in 3 Silene species. A cytological approach. Cytologia. 76(4): 395–402.
- Sosa MM, Panseri AF, Fernández A. 2011. Karyotype analysis of the southernmost South American species of Stemodia (Scrophulariaceae). Plant Biosyst. 145: 472–477.
- Tanaka N, Uchiyama H, Matoba H, Koyama T. 2009. Karyological analysis of the genus Canna (Cannaceae). Plant Syst Evol. 280(1–2): 45–51.
- Tarinejada AR, Mirshekari B. 2010. Study on variation of karyotypes between and within species of Salvia officinalis L., Stachys byzantine L. and Dracocephalum molaricaurae L. Acta Horticult. 853(28): 39–46.
- Uslu E. 2012. Karyology of nine Trifolium L. taxa from Turkey. Caryologia. 65(4): 304–310.
- Vega AJ, Dematteis M. 2012. Chromosome studies of some species of Vernonanthura and Vernonia (Asteraceae, Vernonieae). Caryologia. 65(4): 271–275.
- Viciani D, Fiorini G, Gonnelli V, Gottschlich G. 2013. Karyological and morphological investigations on a Hieracium putatively endemic to the National Park “Foreste Casentinesi, M. Falterona, Campigna” (northern Apennines, central Italy). Caryologia. 66(2): 154–161.
- Wang X, Liu B-B, Ma Y-Z, Xie P-H, He X-Y, Shang B-L, Wang Y-J. 2013. Chromosomal studies on the alpine genus Dolomiaea (Asteraceae: Cardueae) from the Qinghai-Tibet Plateau and adjacent regions. Caryologia. 66(2): 186–193.
- White J, Rees H. 1987. Chromosome weights and measures in Petunia. Heredity. 58: 139–143.
- Zhang Y, Wu M, Yi H, Zhang Y, Zhai W, Feng J. 2010. Studies on mitotic chromosome preparation and karyotype analysis of two different groups of melon in China. Acta Horticult. 871(31): 665–672.
- Zhang Y, Zhang Ch, Zhang T, Guang H, Yan S. 2010. A cyto-evolutional study of Campanumoea Blume (Campanulaceae) and a possible pathway for secondary karyotype formation. Plant Syst Evol. 285(3): 245–257.
- Zhang Y, Zhang Ch, Zhang T, Guang H. 2011. Karyotypic studies on Campanumoea (Campanulaceae) – endemic to China. Genet Resour Crop Evol. 58(3): 461–470.
- Zhou Y, Zhang D, Lu L, Luo Y, Bian L. 2011. Observation and karyotype analysis of chromosomes in somatic cells of root-tips of Chongming narcissus (Narcissus tazetta var. Chinensis). J Plant Resour Environ. 20(2): 56–62.