Abstract
A karyotypic study on South American representatives of the Morelloid and Dulcamaroid clades of Solanum was conducted to contribute to a better understanding of their relationships. Mitotic chromosomes of 26 species were examined (13 of which were previously unknown). All taxa presented 2n = 2x = 24 and had small chromosomes (less than 4 μm long) with the exception of S. crispum. Most species displayed symmetrical karyotypes, being 78% and 69% m, 19% and 25% sm and 3% and 6% st for the Morelloid and Dulcamaroid clades, respectively. Solanum crispum (Dulcamaroid clade) was unique by having mostly sm chromosomes and S. sinuatirecurvum (Morelloid clade) stood out with four st pairs and an exclusive pair of satellites in long arms. Solanum tripartitum was the sole entity exhibiting an sm pair with satellites. Most examined species of these clades resulted karyologically indistinguishable, based on conventionally stained mitotic chromosomes. Molecular analyses are needed to gain a better knowledge of the possible karyoevolutionary trends of both clades.
Introduction
Solanum L. is the largest genus of Solanaceae, with around 1400 species (Bohs Citation2005). It includes plants of economic significance: for their edible tubers, fruits or leaves, for their pharmaceutical compounds or for their ornamental value (Hunziker Citation2001; Weese and Bohs Citation2007); in addition, some are weeds of importance (Hunziker Citation2001).
Systematically, it is a complex genus. The classification of D’Arcy (Citation1991) recognized seven subgenera, whereas other botanists made other infrageneric schemes based on different morphological characters (Nee Citation1999; Child and Lester 2001; Hunziker Citation2001). More recently, phylogenies have been produced and a natural infrageneric arrangement has been proposed (Olmstead et al. Citation1999, Citation2008; Bohs Citation2005; Weese and Bohs Citation2007), with 13 main clades including Morelloid and Dulcamaroid. These clades are closely related and are considered a sister group of ancient origin (Bohs Citation2005; Weese and Bohs Citation2007). Their monophyly was proposed according to sequences of ndhF chloroplast genes, DNA restriction sites, and nuclear ITS DNA sequence analyses (Olmstead and Palmer Citation1997; Bohs and Olmstead Citation1999; Bohs Citation2005; Weese and Bohs Citation2007).
The Morelloid clade has a worldwide distribution but predominates in the New World (Weese and Bohs Citation2007; Barboza et al. Citation2013), with South America being a conspicuous region due to the number of species. This clade mostly agrees with Solanum section Morella (Dunal) Bitter, which includes weeds and food plants. After Bohs (Citation2005), its morphological synapomorphies include: vining habit in many taxa, presence of unbranched, dendritic, or echinoid hairs, 3- to many-foliate sympodial units, and fruits without stone cell aggregates. The Dulcamaroid clade is also distributed worldwide (Eurasia and the Americas) with its centre of diversity in the Andes and south-eastern Brazil (Knapp Citation2013); it includes entities from D’Arcy’s three subgenera (Citation1991). Some morphological features of this clade are: herbaceous or weakly woody habit, 2- to 3-foliate sympodial units, pubescent filaments and styles in many taxa, and small stone cell aggregates in the fruits (Bohs Citation2005).
The knowledge of the structural and quantitative characteristics of the karyotype has proven to be important in evolutionary and taxonomic studies in several angiosperm groups (Shan et al. Citation2003; Weiss-Schneeweiss et al. Citation2003; Urdampilleta et al. Citation2005) and also in Solanaceae (e.g. Fregonezi et al. Citation2006; Moscone et al. Citation2007). There are numerous cytogenetic analyses dealing with Solanum species of economic importance performed with classical and molecular techniques (e.g. Tanskley et al. Citation1992; Dong et al. Citation2000; Cheema and Pant Citation2013). However, the karyology of less than half of Solanum species has been studied (Hunziker Citation2001). Karyotypes have been reported for several American species of Solanum and have proved to be useful in differentiating taxa and evolutionary trends; for instance, Basarthrum (Bernardello and Anderson Citation1990), Lasiocarpa (Bernardello et al. Citation1994), Leptostemonum (Chiarini and Bernardello Citation2006), and Solanum as a whole (e.g. Acosta et al. Citation2005, Citation2012; Rego et al. Citation2009; Chiarini et al. Citation2010; Melo et al. Citation2011).
In the Morelloid and Dulcamaroid clades, few karyotype studies have been conducted (Moscone Citation1992; Acosta et al. Citation2005, Citation2012). Thus, in the present work, we provide a detailed morphometric karyotype analysis for 26 species, 13 of which are here reported for the first time. Our aims are: (1) to describe the chromosome number and to determine the karyotypes; and (2) to contribute to the improvement of the classification system and karyotype evolutionary trends in the Morelloid and Dulcamaroid clades.
Materials and methods
The species studied, locations, and collectors are listed in Table . Vouchers are kept at the herbarium of the Museo Botánico de Córdoba (CORD).
Table 1. Collection data of material studied of the Morelloid and Dulcamaroid clades, indicating previous reports when available. Asterisks indicate the first count and karyotype for the species.
Mitotic chromosomes were examined in squashes of root tips obtained from germinating seeds. Seeds were soaked for 24 h in running water and then put in Petri dishes on moist filter paper at 25 °C. Root tips were pretreated with 2 mM 8-hydroxyquinoline for 6 h at 14 °C, fixed in ethanol : acetic acid (3:1, v/v) for 24 hours and stored at –20 °C until use for conventional chromosome analysis according to the HCl/Giemsa technique (Guerra Citation1983). At least five metaphases of each species were analysed with a Zeiss Axiophot phase contrast microscope (Jena, Germany) and photographed with a Leica DFC300FX camera (Wetzlar, Germany). Photographs were used to take the following measurements for each chromosome pair: s (short arm), l (long arm) and c (total chromosome length), using the freeware computer application MicroMeasure version 3.2 available on the internet at http://www.colostate.edu/Depts/Biology/MicroMeasure. The arm ratio (r = l/s) was then calculated and used to classify the chromosomes as defined by Levan et al. (Citation1964). Karyotype asymmetry was obtained using the intrachromosomal (A1) and the interchromosomal (A2) asymmetry index described by Romero Zarco (Citation1986). Chromosome types were classified first into groups according to their increasing arm ratio and then according to the decreasing length within each group. To look for associations between pairs of the mentioned karyotype variables, linear regression tests were performed using Infostat (Infostat Group Citation2002).
Results
Karyotype analysis of the 26 Solanum species here studied showed a constant chromosome number: 2n = 2x = 24. The chromosome numbers of 13 species, indicated with an asterisk in Table , are reported for the first time.
Table summarizes the morphometric characteristics of each taxa. The Morelloid clade has small chromosomes. The largest chromosome length and haploid genome length values were observed in S. aloysiifolium 2210 (2.07 μm and 24.79 μm, respectively), whereas the lowest ones were detected in S. palitans (0.97 μm and 11.68 μm, respectively). Solanum crispum from the Dulcamaroid clade was outstanding, not only between both clades but also in the genus, for having the largest chromosome length and haploid genome length (4.09 μm and 49.15 μm, respectively). On the other hand, also within the Dulcamaroid clade, S. salicifolium 3158 presented the lowest chromosome length (1.23 μm), whereas the lowest haploid genome lengths corresponded to S. aligerum (18.53 μm).
Table 2. The Solanum taxa studied of the Morelloid and Dulcamaroid clades, with karyotype formulae, total haploid genome length (tl), mean chromosome length (C), mean arm ratio (r), mean asymmetry indices (intrachromosomic: A1, interchromosomic: A2), category of asymmetry of Stebbins (St), and standard deviation (SD).
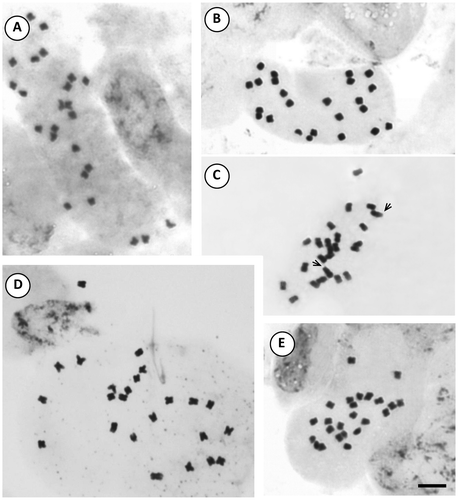
The presence of satellites was inconsistent in the examined species, varying according to the sample. The number of cells in which they could be visualized was variable and satellites could be detected only in one homologue. The occurrence of one satellited pair is common in the Morelloid clade: seven species displayed it in the short arms, and S. pilcomayense 2287 in the long arms (Figures , ). Six analysed species of the Dulcamaroid clade exhibited one pair of satellites in the short arms. Solanum crispum was also outstanding because its satellites were larger than the short arms in which they were attached (Figures , ).
Figure 4. Idiograms of Solanum species of Dulcamaroid clade. (A) S. aligerum; (B) S. amygdalifolium; (C) S. angustifidum; (D) S. crispum; (E) S. dulcamara. Scale bar = 3 μm.
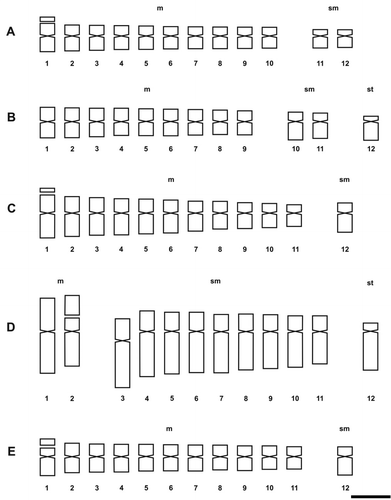
Figure 3. Photomicrographs of mitotic metaphases in Solanum species of Dulcamaroid clade with 2n = 24. (A) S. aligerum; (B) S. amygdalifolium; (C) S. angustifidum; (D) S. crispum; (E) S. dulcamara. Scale bar = 6 μm, all photomicrographs at the same scale. Arrows indicate satellites.
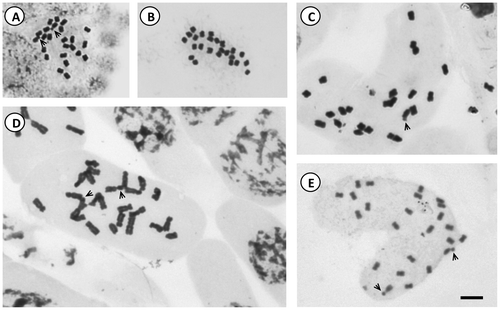
Figure 2. Idiograms of Solanum species of Morelloid clade. (A) S. hastatilobum; (B) S. palitans; (C) S. pilcomayense 2287; (D) S. pilcomayense 2279; (E) S. reductum; (F) S. sarrachoides. Scale bar = 3 μm.
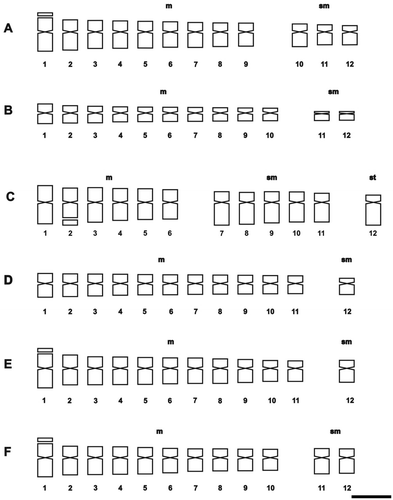
Figure 1. Photomicrographs of mitotic metaphases in Solanum species of Morelloid clade with 2n = 24. (A) S. echegarayi; (B) S. palitans; (C) S. pilcomayense 2279; (D) S. pilcomayense 2287; (E) S. reductum; (F) S. sarrachoides. Scale bar = 6 μm, all photomicrographs at the same scale. Arrows indicate satellites.
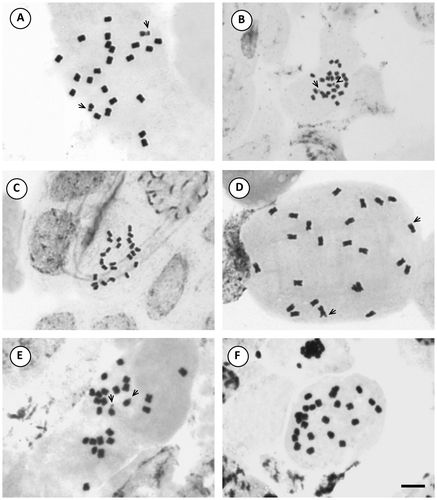
The more frequent karyotype formula of the Morelloid clade was 10m + 2sm (in S. aloysiifolium 2210, S. americanum, S. chenopodioides, S palitans, S. sarrachoides, S. triflorum, S. tripartitum), followed by 11m + 1sm (S. concarense, S. pilcomayense 2279, S. reductum, S. zuloagae), and 9m + 3sm (S. echegarayi, S. hastatilobum) (Figures , , , , ). Three species displayed a unique formula showing st pairs: S. pilcomayense 2287 with 6m + 5sm + 1st, S. aloysiifolium 2152 with 7m + 4 sm + 1st, and S. sinuatirecurvum with 5m + 3sm + 4st (Figures , , ).
Figure 6. Photomicrographs of mitotic metaphases in Solanum species of Morelloid clade with 2n = 24. (A) S. sinuatirecurvum; (B) S. triflorum; (C) S. tripartitum; (D) S. zuloagae. Scale bar = 6 μm, all photomicrographs at the same scale. Arrows indicate satellites.
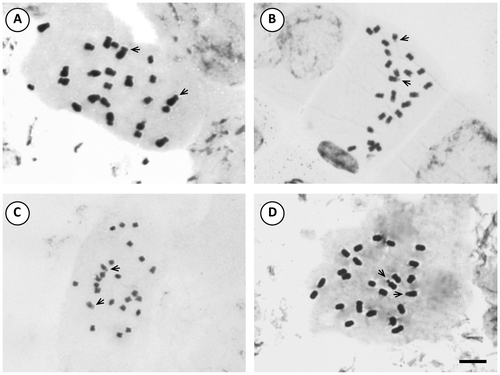
Figure 5. Photomicrographs of mitotic metaphases in Solanum species of Morelloid clade with 2n = 24. (A) S. aloysiifolium 2210; (B) S. aloysiifolium 2152; (C) S. americanum; (D) S. chenopodioides; (E) S. concarense; (F) S. hastatilobum. Scale bar = 6 μm, all photomicrographs at the same scale. Arrows indicate satellites.
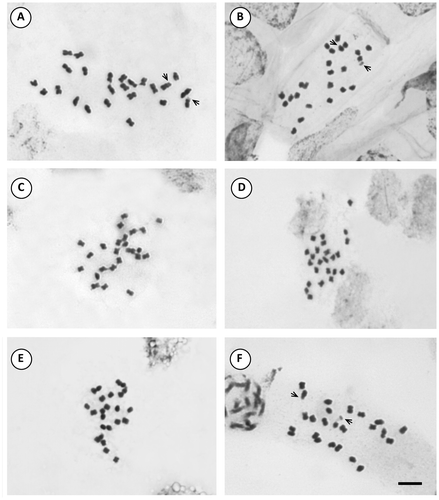
In the Dulcamaroid clade, the majority of species exhibit m chromosomes (Figures , , , ), except in S. crispum with nearly all sm chromosomes (Figure ). S. amygdalifolium showed st chromosomes, along with S. salicifolium 3488 (9m + 2sm + 1st) and S. salicifolium 3158 (7m + 4sm + 1 st). In contrast, S. aligerum, S. salicifolium 818, S. salicifolium 794 (10m + 2 sm), S. angustifidum and S. dulcamara (11m + 1sm) (Figures , C, E, , E) did not show st chromosomes. Neither in this clade nor in the Morelloid clade were t chromosomes detected.
Figure 10. Idiograms of Solanum species of Morelloid clade. (A) S. sinuatirecurvum; (B) S. triflorum; (C) S. tripartitum; (D) S. zuloagae. Scale bar = 3 μm.
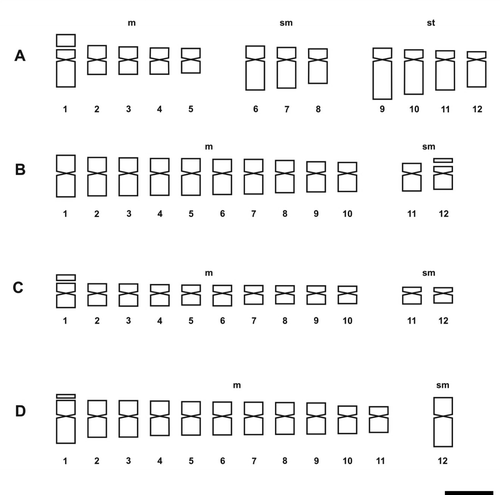
Figure 9. Idiograms of Solanum species of Morelloid clade. (A) S. aloysiifolium 2210; (B) S. aloysiifolium 2152; (C) S. americanum; (D) S. chenopodioides; (E) S. concarense; (F) S. echegarayi. Scale bar = 3 μm.
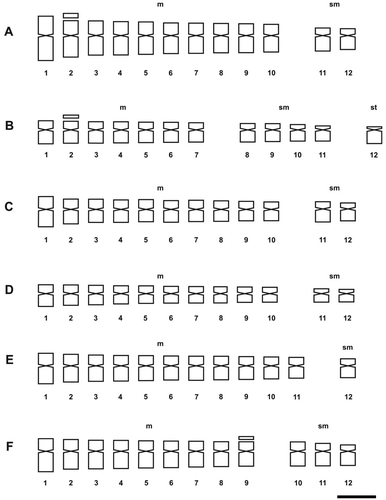
Figure 8. Idiograms of Solanum species of Dulcamaroid clade. (A) S. endoadenium; (B) S. salicifolium 3488; (C) S. salicifolium 3158; (D) S. salicifolium 818; (E) S. salicifolium 794. Scale bar = 3 μm.
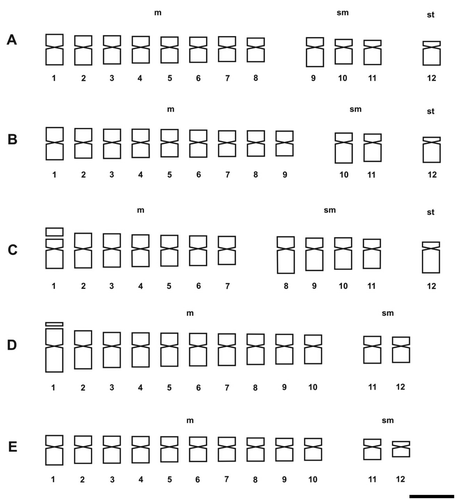
Figure 7. Photomicrographs of mitotic metaphases in Solanum species of Dulcamaroid clade with 2n = 24. (A) S. endoadenium; (B) S. salicifolium 3488; (C) S. salicifolium 818; (D) S. salicifolium 3158; (E) S. salicifolium 794. Scale bar = 6 μm, all photomicrographs at the same scale. Arrows indicate satellites.
The karyotypes of the studied taxa were mainly symmetrical; S. pilcomayense 2287 with A1 + A2 = 0.35 (Morelloid clade) and S. amygdalifolium with A1 + A2 = 0.30 (Dulcamaroid clade) were the most symmetrical (Table ). S. sinuatirecurvum with A1 + A2 = 0.67 (Morelloid clade) and S. crispum with A1 + A2 = 0.64 (Dulcamaroid clade) were the most asymmetrical. After the classification of Stebbins (Citation1971), the species of both clades showed different categories. Nine Morelloid clade species fell into the 1A category, four into 1B, two into 2B and just S. sinuatirecurvum into 2C. In the Dulcamaroid clade, three species fell into the 1A category, three into 1B, two into 2B, and one into each of 2A and 1C (S. angustifidum and S. crispum, respectively).
Discussion
Solanum species here examined are diploid with x = 12. This is the most frequent number for the genus and for Solanaceae as well, being present in more than 50% of the known species (cf. Acosta and Moscone Citation2000; Hunziker Citation2001; Acosta et al. Citation2005, Citation2012). A few exceptions have been detected in Solanum within the Leptostemonum clade (S. mamosum and S. platense with n = 22; Chiarini and Bernardello Citation2006) and the Archaesolanum clade (seven species with n = 23; Pinto Maggio et al. Citation1997).
Solanum as a whole shows small chromosomes of less than 4 μm (e.g. Bernardello and Anderson Citation1990; Bernardello et al. Citation1994; Acosta et al. Citation2005; Chiarini and Bernardello Citation2006; Rego et al. Citation2009; Melo et al. Citation2011), as mostly found here. The Cyphomandra clade is an exception with chromosome sizes ranging from 3 to 14 μm (e.g. Pringle and Murray Citation1991; Moscone Citation1992; Miguel et al. 2012), as found here in S. crispum with chromosomes longer than 4 μm.
In Solanaceae and in Solanum there is a prevalence of m and sm chromosomes (e.g. Trivedi and Sinha Citation1986; Bernardello and Anderson Citation1990; Bernardello et al. Citation1994; Moscone Citation1990, 1999; Acosta and Moscone Citation2000; Acosta et al. Citation2005; Chiarini and Bernardello Citation2006). On the other hand, st chromosomes in Solanum are rare, either as one pair (e.g. Trivedi and Sinha Citation1986; Bernardello and Anderson Citation1990; Acosta et al. Citation2005; Chiarini and Bernardello Citation2006) or two pairs (e.g. Bernardello et al. Citation1994; Chiarini and Bernardello Citation2006). In the Morelloid clade, we detected an outstanding species with four st pairs: S. sinuatirecurvum. This is the first report in this clade, and even in the genus Solanum, with this many st pairs.
The number of satellited pairs is variable within Solanaceae; for instance, Capsicum species displayed one to four satellited pairs (Moscone 1999), Lycium always showed one (Stiefkens and Bernardello Citation2002) and species of Hyoscyamus may have one to three pairs (Sheidai et al. Citation1999). Many Solanum showed satellites in the short arms: one pair (as here found in both clades), two pairs (in S. pseudolulo, Bernardello et al. Citation1994), three pairs (in species of the S. indicum complex, Krishnappa and Chennaveeraiah 1975). There are relatively few reported cases in which satellites were located on the long arms, namely Nicotiana (Moscone 1989), Datura (Palomino and Bye 1988; Moscone 1989), Cestrum (Sharma and Sharma Citation1957), and Solanum (Bernardello and Anderson Citation1990). Solanum pilcomayense 2287 (Morelloid clade) was the only taxon studied with satellites in the long arms. Regarding the magnitude of the satellites, it can vary depending on the species, being of remarkable size in S. crispum (Dulcamaroid clade), where they are even larger than the arms.
The genus Solanum is complex and there are many problems in delimiting its taxa and defining an acceptable infrageneric circumscription (e.g. D’Arcy Citation1991; Nee Citation1999; Bohs Citation2005; Weese and Bohs Citation2007). Our karyotypic analysis of species of the Morelloid and Dulcamaroid clades allowed the detection of differences between the examined taxa. Thus, chromosome variation, although not always high, accompanied evolutionary divergence of the species studied, a general phenomenon observed in both plant and animal kingdoms (e.g. Goodspeed Citation1954; Rieseberg Citation2001). At the same time, karyotype similarities may indicate relationships between taxa. Based on morphological features, Nee (Citation1999), does not consider S. triflorum, S. tripartitum and S. palitans to be closely related species, placing S. triflorum in sect. Solanum (Morelloid clade) and S. tripartitum and S. palitans in sect. Dulcamara (Dulcamaroid clade). By contrast, Bohs (Citation2005) and Weese and Bohs (Citation2007), analysing sequences of the ndhF chloroplast genes, and Barboza et al. (Citation2013), analysing morphology, included them in the Morelloid clade. Our results agreed with the latter scheme, since these taxa displayed the same karyotypic formula. At the same time, S. tripartitum and S. palitans fell in the category 1A of Stebbins (Citation1971) and showed similar values of tl and C (Table ), whereas S. triflorum was included in the 2B category and its values of tl and C were higher. These facts would indicate that S. tripartitum and S. palitans are closely related species, in agreement with Bohs (Citation2005) and Weese and Bohs (Citation2007). Moreover, these two species are morphologically very similar, to the point that they are often confused (Barboza et al. Citation2013).
Stebbins (Citation1971) included Solanum in his 1A category due to the known presence of highly symmetrical karyotypes, as was later reported by several authors (e.g. Bernardello and Anderson Citation1990; Acosta et al. Citation2005; Chiarini and Bernardello Citation2006). However, our data indicated that the Morelloid and Dulcamaroid clades have a range of karyotypes, from symmetrical (most species) to less expectedly, more asymmetrical (e.g. S. sinuatirecurvum, S. crispum).
Recent data indicate that Solanaceae in general, and Solanum in particular, have a low rate of chromosomal changes and that many small rearrangements not observed at the cytological level have occurred (Wu and Tanskley Citation2010), as previously suggested (e.g. Stebbins Citation1971; Bernardello and Anderson Citation1990; Bernardello et al. Citation1994).
Most examined species of these clades were karyologically indistinguishable, based on conventionally stained mitotic chromosomes, although some species are clearly noticeable, such as S. sinuatirecurvum and S. crispum. Additional molecular karyotypic analyses with banding and FISH techniques are needed in the Morelloid and Dulcamaroid clades to gain a better knowledge of the possible karyoevolutionary trends.
Acknowledgements
We are grateful to G. E. Barboza and F. E. Chiarini for specimen identification, to Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) and Secretaría de Ciencia y Tecnología (Universidad Nacional de Córdoba) for financial support, and to the anonymous reviewers for helpful comments.
References
- Acosta MC, Bernardello G, Guerra M, Moscone EA. 2005. Karyotype analysis in several South American species of Solanum and Lycianthes rantonnei (Solanaceae). Taxon. 54(3):713–723.
- Acosta MC, Guerra M, Moscone EA. 2012. Karyological relationships among some South American species of Solanum (Solanaceae) based on fluorochrome banding and nuclear DNA amount. Plant Syst Evol. 298(8):1547–1556.
- Acosta MC, Moscone EA. 2000. Estudio cariotípico en Dyssochroma viridiflorum (Solanaceae). Bol Soc Argent Bot. 35(3–4):227–236.
- Barboza GE, Knapp S, Särkinen TE. 2013. Solanaceae. In: Anton AM, Zuloaga FO, editors. Flora Argentina. vol 13. Buenos Aires: IBODA-IMBIV, 231–264.
- Bernardello L, Anderson GJ. 1990. Karyotypic studies in Solanum section Basarthrum (Solanaceae). Am J Bot. 77(3):420–431.
- Bernardello L, Heiser CB, Piazzano M. 1994. Karyotypic studies in Solanum sect. Lasiocarpa (Solanaceae). Am J Bot. 81(1):95–103.
- Bir S, Kumari S, Shoree S, Sagoo M. 1978. Cytological studies in certain Bicarpellate from north and central India. J Cytol Genet. 13:99–106.
- Bohs L. 2005. Major clades in Solanum based on ndhF sequence data. Monogr Syst Bot Missouri Bot Gard. 104:27–50.
- Bohs L, Olmstead R. 1999. Solanum phylogeny inferred from chloroplast DNA sequence data. In: Nee M, Symon DE, Lester RN, Jessop JP, editors. Solanaceae IV: advances in biology and utilization. Kew: Royal Botanical Gardens, 97–110.
- Cheema SK, Pant MR. 2013. Karyotype analysis of seven cultivated varieties of Capsicum annuum L. Caryologia. 66(1):70–75.
- Chiarini F, Bernardello G. 2006. Karyotype studies in South American species of Solanum subgen. Leptostemonum (Solanaceae). Plant Biol. 8(4):486–493.
- Chiarini FE, Moreno NC, Barboza GE, Bernardello G. 2010. Karyotype characterization of Andean Solanoideae (Solanaceae). Caryologia. 63(3):278–291.
- Child A, Lester, RN. 2001. Synopsis of the genus Solanum L. and its infrageneric taxa. In: van den Berg RG, Barendse GWM, van der Weerden GM, Mariani C., editors. Solanaceae V: advances in taxonomy and utilization. Nijmegen: Nijmegen University Press. p. 39–52.
- D’Arcy WG. 1991. The Solanaceae since 1976, with a review of its biogeography. In: Hawkes JG, Lester R, Nee M, Estrada N, editors. Solanaceae III: Taxonomy, Chemistry, Evolution. Kew: Royal Botanic Gardens, 75–137.
- Dong F, Song J, Naess SK, Helgeson JP, Gebhardt C. 2000. Development and applications of a set of chromosome specific cytogenetic DNA markers in potato. Theor Appl Genet. 101:1001–1007.
- Fregonezi JN, Fernandes T, Dominguez Torezan JM, Vieira O, Vanzela ALL. 2006. Karyotype diffentiation of four Cestrum species (Solanaceae) based on physical mapping of repetitive DNA. Genet Mol Biol. 29(1):97–104.
- Ganapathi A, Rao R. 1982. Interrelationships between tetraploid Solanum nigrum L. and Solanum americanum Mill. Proc Indian Sci Congr Assoc. 69:224.
- Goodspeed TH. 1954. The Genus Nicotiana. Waltham: Chronica Botanica.
- Guerra MS. 1983. O uso do Giemsa em Citogenetica Vegetal: comparação entre a coloração simples e o bandamento. Ciênc Cultura. 35:190–193.
- Hunziker AT. 2001. Genera Solanacearum. Ruggell: ARG Gantner Verlag KG.
- Infostat Group. 2002. INFOSTAT. Version 1.1. Córdoba: Facultad de Ciencias Agropecuarias, Universidad Nacional de Córdoba.
- Knapp S. 2013. A revision of the Dulcamaroid clade of Solanum L. (Solanaceae). PhytoKeys. 22:1–432.
- Krishnappa DG, Chennaveraiah MS. 1975. Cytotaxonomy of Solanum indicum complex. Cytologia. 40:323–331.
- Levan A, Fredga L, Sandberg A. 1964. Nomenclature for centromeric position on chromosomes. Hereditas. 52:201–220.
- Melo CAF, Martins MIG, Oliveira MBM, Benko-Iseppon AM, Carvalho R. 2011. Karyotype analysis for diploid and polyploid species of the Solanum L. Plant Syst Evol. 293(1–4):227–235.
- Miguel V, Acosta MC, Moscone EA. 2012. Karyotype analysis in two species of Solanum (Solanaceae) sect. Cyphomandropsis based on chromosome banding. New Zeal J Bot. 50(2):217–225.
- Moscone EA. 1989. Karyotype analysis in three Patagonian and S. Andean endemic genera of Nicotianeae (Solanaceae). Plant Syst Evol. 166(1–2):31–39.
- Moscone EA. 1990. Chromosome studies on Capsicum (Solanaceae) I. Karyotype analysis in C. chacöense. Brittonia. 42:147–154.
- Moscone EA. 1992. Estudios sobre cromosomas meióticos en Solanaceae de Argentina. Darwiniana. 31(1–4):261–297.
- Moscone EA. 1999. Análisis cariotípico en Capsicum baccatum var. umbilicatum (Solanaceae) mediante bandeos AgNOR y de fluorescencia. Kurtziana. 27(1):225–232.
- Moscone EA, Scaldaferro MA, Grabiele M, Cecchini NM, Sánchez García Y, Jarret R, Daviña JR, Ducasse DA, Barboza GE, Ehrendorfer F. 2007. The evolution of chili peppers (Capsicum - Solanaceae): a cytogenetic perspective. Acta Horticulturae. 745:137–169
- Nee M. 1999. Synopsis of Solanum in the New World. In: Nee M, Symon DE, Lester RN, Jessop JP, editors. Solanaceae IV: Advances in Biology and Utilization. Kew: Royal Botanic Gardens, 285–333.
- Olmstead RG, Bohs L, Migid HA, Santiago-Valentin E, García VF, Collier SM. 2008. A molecular phylogeny of the Solanaceae. Taxon. 57(4):1159–1181.
- Olmstead RG, Palmer JD. 1997. Implications for the phylogeny, classification, and biogeography of Solanum from cpDNA restriction site variation. Syst Bot. 22(1):19–29.
- Olmstead RG, Sweere JA, Spangler RE, Bohs L, Palmer JD. 1999. Phylogeny and provisional classification of the Solanaceae based on chloroplast DNA. In: Symon DE, Lester RN, Jessop JP, editors. Nee M. Royal Botanic Gardens: Solanaceae IV. Advances in Biology and Utilization. Kew, 257–274.
- Palomino GR, Bye RA. 1988. Cytology of five Mexican species of Datura L. (Solanaceae). Southwest Nat. 33(1):85–90.
- Pinto Maggio CAF, Pierozzi NL, Castro SCP, Soares Scott MD. 1997. Solanaceae. In: Stace CA, editor. IOPB Chromosome Data, 11–24.
- Pringle GJ, Murray BG. 1991. Karyotypes and G-banding patterns in species of Cyphomandra Mart. Ex Sendtn. (Solanaceae). Bot J Linn Soc. 111:331–342.
- Rego LNA, da Silva CRM, Torezan JMD, Gaeta ML, Vanzela ALL. 2009. Cytotaxonomical study in Brazilian species of Solanum, Lycianthes and Vassobia (Solanaceae). Plant Syst Evol. 279(1):93–102.
- Rieseberg LH. 2001. Chromosomal rearrangements and speciation. Trends Ecol Evol. 16(7):351–358.
- Romero Zarco C. 1986. A new method for estimating karyotype asymmetry. Taxon. 35(3):556–530.
- Shan F, Yan G, Plummer JA. 2003. Karyotype evolution in the genus Boronia (Rutaceae). Bot J Linn Soc. 142(3):309–320.
- Sharma AK, Sharma A. 1957. Karyotype studies in Cestrum as an aid to taxonomy. Genetica. 29(1):83–100.
- Sheidai M, Mosallanejad M, Khatamsaz M. 1999. Karyological studies in Hyoscyamus species of Iran. Nord J Bot. 19(3):369–373.
- Stebbins GL. 1971. Chromosomal evolution in higher plants. London: E. Arnold.
- Stiefkens L, Bernardello G. 2002. Karyotypic studies in Lycium section Mesocope (Solanaceae) from South America. Caryologia. 55(3):199–206.
- Tanskley SD, Ganal MW, Prince JP, de Vicente MC, Bonierbale MW, Broun P, Fulton TM, Giovannoni JJ, Grandillo S, Martin GB, et al. 1992. High density molecular linkage maps of the tomato and potato genomes. Genetics. 132(4):1141–1160.
- Trivedi RN, Sinha AK. 1986. Karyomorphological studies in three population of Solanum suruttense, a weed. Cytologia. 51:157–161.
- Urdampilleta JD, Ferrucci MS, Vanzela ALL. 2005. Karyotype differentiation between Koelreuteria bipinnata and K. elegans ssp. formosana (Sapindaceae) based on chromosome banding patterns. Bot J Linn Soc. 149(4):451–455.
- Weese TL, Bohs L. 2007. A three–gene phylogeny of the genus Solanum (Solanaceae). Syst Bot. 32(2):445–463.
- Weiss-Schneeweiss H, Stuessy TF, Siljak-Yakovlev S, Baeza CM, Parker J. 2003. Karyotype evolution in South American species of Hypochaeris (Asteraceae, Lactuceae). Plant Syst Evol. 241(3–4):171–184.
- Wu F, Tanskley SD. 2010. Chromosomal evolution in the plant family Solanaceae. BMC Genomics. 11:182.