Abstract
The amount of DNA in the unreplicated haploid nuclear genome (C-value) provides important information for genome biodiversity. Genome size is used in a strikingly wide variety of plant biological fields with both practical and biological significance. To extend previously published datasets on plant nuclear content and to compile information on the DNA content of traditional Chinese medicinal plants in China, flow cytometry (FCM) was used to estimate C-values of medicinal plants in Shaanxi province, China. A total of 80 medicinal plant species and vouchers were collected from locations across Shaanxi province in China. Nuclei were extracted from fresh leaves in one of two buffers, stained with fluorochrome propidium iodide (PI), and analyzed on a Guava flow cytometer to measure the position of the fluorescence peaks relative to those of an internal calibration standard. Replicate extractions, low coefficients of variation and comparisons with published C-values in the same and related species were used to confirm the accuracy and reliability of our results. Prime C-values for 66 medicinal plant taxa are provided for which no published data exist, comprising 66 angiosperms, one gymnosperm and one pteridophyte. C-values for 14 additional taxa where a genome size has previously been published are also provided. The prime values represent new reports for 27 genera (out of the 48 genera and 38 families sampled). These data provide the foundation to enable phylogenetic analysis of C-value variation and karyotype diversity in Chinese traditional medicinal plants and assist future analyses aimed at analyzing how C-values co-vary with effective components and functional traits in Chinese traditional medicinal plants.
Introduction
Genome sizes are characterized in terms of their C-value, the picograms of DNA present in an unreplicated haploid or gametic nucleus (Swift Citation1950). The size of an organism’s genome reflects a fundamental aspect of its biology as well as a character of considerable practical use (Fay et al. Citation2005; Bennett and Leitch Citation2005, Citation2011; Beaulieu, Leitch, et al. Citation2007; Beaulieu, Moles, et al. Citation2007; Beaulieu et al. Citation2008; Leitch et al. Citation2009, Citation2010; Chung et al. Citation2012; Leitch and Leitch Citation2012; Janousek et al. Citation2013). To date, researchers have estimated C-values for 8509 plant species, as reported in the most recent release of the Plant DNA C-values Database (Release 6.0): http://data.kew.org/cvalues/ (Bennett and Leitch Citation2012). The electronic databases have now been cited > 230 times with > 250,000 hits (Bennett and Leitch Citation2011), and provide a powerful global research platform for biological scientists across the world. Researchers have used DNA C-values to address questions in cellular, developmental, ecological, evolutionary and molecular biology as well as systematics, physiology and paleontology (e.g. Bennett et al. Citation2000; Bennett and Leitch Citation2005; Leitch and Bennett Citation2007; Franks et al. Citation2012; Greilhuber and Leitch Citation2013). Recent work includes studies of the relationships between genome size and seed mass (Beaulieu, Moles, et al. Citation2007), photosynthetic rate (Beaulieu, Leitch, et al. Citation2007), leaf cell size and stomata density (Beaulieu et al. Citation2008; Hodgson et al. Citation2010) and patterns of genome size evolution (Leitch et al. Citation2005, Citation2009; Beaulieu et al. Citation2010; Chung et al. Citation2012). With the increase in the number of plant C-values in the Plant DNA C-values Database, studies of the relationship between genome size and various biological characteristics is likely to continue to expand.
Over the years, flow cytometry and Feulgen microdensitometry have both been used extensively to estimate C-values in plants, although flow cytometry is becoming increasingly the method of choice as the machinery needed for Feulgen microdensitometry becomes less widely available (Bennett and Leitch Citation2011). Nevertheless, flow cytometry is not without its problems and careful application of the technique is essential for generating reliable data (Doležel et al. Citation2007a; Greilhuber et al. Citation2007). Indeed, there are multiple factors that can affect the reliability of C-value data, including the choice of extraction buffer (with over 28 different buffers in use; Lourerio et al. Citation2007), how raw FCM data are analyzed (forward scatter, side scatter and relative fluorescence intensity), which plant species are used as calibration standards (Doležel and Greilhuber Citation2010; Suda and Leitch Citation2010) and the potentially confounding effects of cytosolic compounds (e.g. anthocyanin and tannic acid) (Doležel et al. Citation2007a, Citation2007b; Bennett and Leitch Citation2011; Loureiro, Suda, et al. Citation2007; Loureiro, Rodriguez, et al. Citation2007; Greilhuber et al. Citation2007).
Traditional Chinese medicine (TCM) is a system of medicine with the longest history in the world and it plays an important role in health maintenance within China and Asia. In recent years TCM has also become increasingly popular in the western world although there have been some problems arising from misunderstandings about what specific plant species are used in TCM and what standards they conform to (Tian Citation2011; Tu Citation2011). TCM has been developed over thousands of years of empirical testing and refinement and the compatibility and quality of the particular medicinal plant or animal species used is critical for its successful application. Indeed, selection of the wrong original medicinal plant in TCM can lead to unpredictable consequences (Xu Citation2011). Though there are lots of plant taxa that contain many of the same main active ingredients in one genus, specific plant species containing the most medicinal ingredients cannot be replaced in TCM. In recent years there has been increasing interest in the genetic diversity, and phylogenetic and evolutionary relationships between the medicinal plants that have been used for TCM (Li et al. Citation2012; Bai et al. Citation2013). One genomic character which has been poorly studied in species used for TCM is genome size, so there is little knowledge about how much variation there is in this character and how it co-varies with phenotypic characters and phylogenetic relationships in the same genus or across the family. Estimating C-values can therefore give us insights into the diversity of this genomic character for medicinal plants and how it varies.
In this study we report C-value estimates for 80 taxa of medicinal plants collected from the medicinal plant germplasm repository or field in Shaanxi province. Most of these reports (83%, 66 taxa) are new to the Plant DNA C-values Database. We had two goals in collecting these data. First, we sought to augment the amount of genome size data available for research generally. More specifically, we sought to start the construction of a database of C-values for Chinese traditional medicinal plant species to explore phylogenetic patterns of C-value variation and relationships between genome size and other qualitative characteristics or functional traits. This work therefore represents the first phase of a broader project to explore how genetic, phylogenetic and qualitative characteristics among medicinal plants vary in same genus or family.
Materials and methods
Plant material
We first identified a list of traditional Chinese medicinal plant species for which we had test data concerning genetic resources and pharmacological active ingredients. We sought to sample fresh leaf material from these species from across the region (Table ). Fresh leaf material was stored at 4°C for no more than three days until it could be processed. For each sample, we collected a herbarium voucher serving as a permanent record (starting with DMP001) of the species identity, and deposited these at the herbarium in Shaanxi Normal University. The taxonomic identities of all field-collected material were confirmed by a medicinal botanist to ensure accurate identifications. We also collected small backup samples of leaf tissue in silica gel. These were used in the few cases where the fresh tissue did not provide a C-value and the DNA was extracted for molecular analysis.
Table 1. Provenance of the 80 plant species collected for this study in Shaanxi province (SN), China.
Estimating DNA C-values
We estimated 2C-values from leaf nuclei using flow cytometry with the fluorochrome propidium iodide (PI, Sigma®, USA, Pcode: 1001159523) as described by Doležel et al. (Citation2007b). We relied primarily on the Otto and LB01 isolation buffers (Doležel et al. Citation1989; Otto Citation1990; Doležel and Gohde Citation1995) using the methods (reagent preparation, selection of standards, etc.) laid out initially by Otto (Citation1990) and available on the flow cytometry methodology webpage (www.ibot.cas.cz/fcm/method.html).
In brief, we placed ~1 cm2 of leaf tissue from both the sample and a known plant calibration standard into a 100 mm diameter Petri dish with 1.0–1.5 ml of ice-cold buffer (Otto I or LB01). We then finely chopped the tissue using a new Blade (Gillette ®, Shanghai, China) for one minute. After mixing this solution using a disposable graduated transfer pipet, we filtered it through a 30 μm disposable nylon filter (Celltrics®, Partec, GmBH, Munster, Germany) into a 1.5 ml round-bottom tube. The filter isolated the somatic nuclei from much of the cellular debris in the isolation buffer. For samples chopped in the Otto I buffer, the filtered suspension was centrifuged at 1000 g for 5 min at 4°C and the supernatant was carefully removed. Then 500 μl of Otto II stock solution (0.02 mg ml−1 RNase + 0.02 mg ml−1 PI) was added at room temperature. For samples chopped in the LB01 buffer 20 μl of the PI stock solution and 20 μl of an RNAase stock solution were added to the nuclei suspension (both stocks were 1 mg ml−1, giving final concentrations of 0.05 mg ml−1). The LB01 or Otto II suspensions were kept on ice. Immediately prior to analysis on the flow cytometer, the nuclei suspensions were vortex-mixed at 2000 rpm for 5–10 s and then placed onto a Guava flow cytometer. Typically, we prepared several samples of each species and analysed > 10,000 nuclei per sample to evaluated the coefficient of variation (where CV = SD/mean channel number, as Ormerod 2008). If the CV was more than 5%, we obtained further estimates until the CV was less than 5%. In general, the two-step procedure of Otto (Citation1990) gave lower CVs and hence more accurate C-value estimates. The most likely reason for this is that the centrifugation of the samples in Otto I buffer removes complex secondary metabolites that can affect nucleus extraction, stability and PI staining. In cases where the Otto buffers did not provide a reliable C-value estimate, we tried using the LB01 buffer to extract nuclei. Overall, we used the Otto and LB01 isolation buffers to estimate C-values in 78 and two species respectively. The 2C-value for each sample was estimated using the following formula: (mean of sample peak/mean of standard peak) × 2C DNA amount (pg) of the calibration standard.
Internal calibration standards
We used three internal standards of different DNA content to calibrate our estimates: Oryza sativa L. subsp. japonica var. nippobare, Pisum sativum var. Bakana, and Vicia faba. We determined the 2C-values of the latter two species in our laboratory (8.32 and 26.57 pg, respectively) using Oryza sativa L. subsp. japonica var. nippobare (2C = 0.91 pg) as the primary reference standard. We grew plants of our three standards in the greenhouse to ensure access to fresh young leaves. To choose the best standard for a given sample, we followed the same protocol as described in Bai et al. (Citation2012). The P. sativum standard, which generated reliable, reproducible, and discernible peaks with low CVs in our pilot tests, was first used to estimate C-values. If it failed, we turned to Oryza sativa and Vicia faba to measure the sample. Since the leaves of the medicinal plants often contain complex secondary metabolites it can be difficult to find the right combination of buffer and internal standard for accurate 2C estimations by FCM. In the few cases where the sample could not be reliably estimated using the approaches outlined above, we abandoned further efforts. In total, we used P. sativum, O. sativa, and V. faba as internal standards to estimate C-values of 55, 24, and one species, respectively.
Results
Accuracy and reliability of the estimated C-values
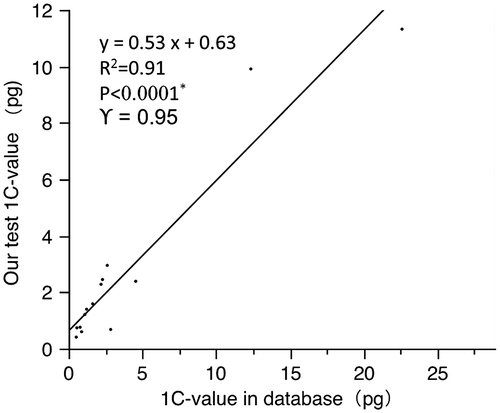
To assess the quality of the data generated, we compared our C-value estimates for 14 species whose C-values had been previously published (see Bennett and Leitch Citation2012; Table ). The overall linear regression analysis showed that there was generally good agreement between the C-values estimated in the current work and those previously published (Figure , R 2 = 0.91, p < 0.0001). For the subset of 10 values (71%) that agreed within 30%, the relationship was even tighter (Figure , R 2 = 0.99, p < 0.0001). In the remaining four species, our estimates differed from previous C-value estimates by > 30%. Though there are differences of the estimation methods and internal calibration standards between our tests and previous references, the C-values of same plant taxa should not have a significant difference. The possible causes of such discrepancies are unclear, but may include intraspecific ploidy and dysploid variation in chromosome numbers, taxonomic discrepancies or genuine intraspecific variation. There were three species where our C-values were half the size or 4× as big as previous estimates. For example, our C-value estimates for Foeniculum vulgare Mill. (1C = 2.39 pg) and Equisetum hyemale L. (1C = 11.32 pg) were half the size of the values in the database (1C = 4.55 pg and 1C = 22.60 pg, respectively) (Bennett et al. Citation2005; Das and Mallick Citation1989) whereas the C-value of Cassia tora L. in the database (1C = 0.68 pg) was just one quarter the size of our estimate 2.85 pg (Ohri et al. Citation1986). We had confidence in our identifications of field-collected plants and all samples are vouchered. Indeed, such variation does highlight the importance of including proper vouchers in all rigorous studies of C-value variation.
Table 2. C-values of 14 species obtained in the current work and those from previous studies.
Figure 1. Comparisons of 1C DNA amounts of 14 taxa tested in this study versus values for the same species taken from the Plant DNA C-values Database (Bennett and Leitch Citation2012).
Reported C-values of Chinese medicinal plants
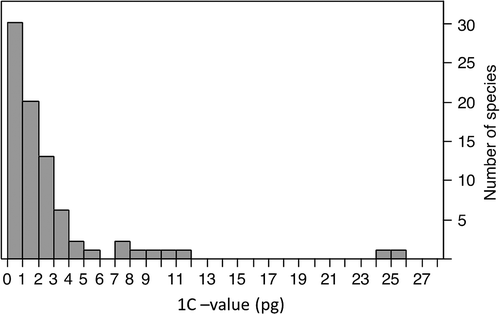
Appendix Table A1 lists the C-values for the 80 Chinese medicinal plant species studied in this work. Of these, 66 (83%) comprise reports for taxa with no previously reported C-value as of release 6.0, December 2012 (Bennett and Leitch Citation2012). The range of 1C-values reported here (0.20–25.2 pg; Figure ) covers a substantial part of the variation in 1C-values reported in the Plant DNA C-values Database (0.0648–152.23 pg) (Greilhuber et al. Citation2006; Pellicer et al. Citation2010). These new C-values represent a valuable addition to the global pool of information on plant genome sizes, with an especially significant addition to knowledge of this character in species used in TCM. The data include several first values at the level of genus. For example, 27 of the 48 genera in 38 families represent new reports at that level for the C-value Plant DNA C-values Database. The C-values of medicinal plant taxa in new genera will give valuable references for estimating other plant C-values and revealing the phylogenetics of total plant taxa.
Discussion
Genome size, as reflected in these plant C-values, represents a key diversity character as well as one that is often associated with other traits such as nuclear and cell size, seed mass, specific leaf area, growth rate and /or cell- and life-cycle length (see Introduction). To better understand the chemical diversity, genetic diversity and phylogenetic relationships of medicinal plant species in the same genus or family, we are also quantifying an array of other attributes for many of the 80 taxa reported here. These include analyses of their karyotypes, chemical composition as well as RNA sequence data for plant taxa in the same genus. Together these data will allow us to gain greater insight into the genomic diversity and phylogenetic relationships among these medicinal plants and compare them with their non-medicinal relatives in a genus or family. This will allow us to analyze how genome size co-varies with these traits. And furthermore, knowledge of a medicinal plant species’ genome size is essential for many genomic studies such as those aiming to embark on complete genome sequencing, transcriptome studies, repetitive DNA characterization, and AFLP and microsatellite analyses for population genetics (Bennett and Leitch Citation2011; Kelly and Leitch Citation2011; Kelly et al. Citation2012; Vallès et al. Citation2013). Although C-value or genome size data are still available for only a small part of the medicinal plants, future analysis will permit us to postulate some hypotheses about systematic, phylogenetic and evolutionary aspects of the genome size (C-value) or chemical diversity of medicinal plants with the data accumulating. For example, we firstly aim to characterize patterns of phenotypic variation, karyotype diversity, genetic diversity by AFLP, chemical composition, phylogenetic relationship and RNA sequences within at least 10 of species of Scutellaria so that we can examine whether genome sizes are associated with such traits. Collectively, these and further studies will extend our ability to understand how genome size varies with phylogenetic, chemical components and genetic diversity.
Appendix. Table A1. C-values of 80 Chinese traditional medicinal plants.
The format of this table is modified from that used by Bennett and colleagues (Bennett and Leitch Citation1997, Citation2011; Bennett et al. Citation2000; Zonneveld et al. Citation2005 – see main text for references). Notes on its structure are as follows.
“New” reports at specific and generic levels. Taxa with C-values first reported here (i.e. for which no previous C-value has been published for this species; Bennett and Leitch Citation2012) are indicated with “+” and appear in bold text; “+ +” indicates that the C-value also is the first report for the genus; “–” indicates that at least one C-value for the taxon has already been published.
Taxa. Names of taxa correspond generally to those provided in the Flora Republicae Popularis Sinicae (http://frps.plantphoto.cn/) and the Directory of Chinese medicinal plants.
Family, order, and class. The family and order assigned to angiosperms correspond with assignments under the Angiosperm Phylogeny Group system (APG III 2009; Stevens Citation2012). Dic = dicotyledoneae; Mon = monocotyledonae; Equ = Equisetopsida; Lep = Leptosporangiopsida; Taxus = Taxopsida.
Life cycle types. Information on the type of life cycle for each taxon was gleaned from the literature or from www.plants.usda.gov/classification.htm: A = annual; B = biennial; P = perennial; AP = annual–perennial (including annual–biennial); BP = biennial–perennial.
DNA amounts and conversion factors. 1C-values were calculated from the available 2C-values. For 1C-values given in megabase pairs (Mbp), we used the conversion factor of 1 pg = 978 Mb (Doležel et al. Citation2003).
Buffers. We relied primarily on the Otto and LB01 isolation buffers (Doležel et al. Citation1989; Otto Citation1990; Doležel and Gohde Citation1995) using the methods (reagent preparation, selection of standards, etc.) laid out initially by Otto (Citation1990) and available on the flow cytometry methodology webpage (www.ibot.cas.cz/fcm/method.html).
Vouchers. This study is part of a larger Diversity of Medicinal Plants (DMB) project at the Shaanxi Normal University (SNNU). To permanently document the taxonomic identity of the source material, and to serve as a reference point for future changes in determinations or nomenclature, we collected herbarium vouchers and deposited these at the herbarium in SNNU.
Acknowledgments
This work was partially funded by Natural Science Foundation of China (No. 31100241), Shaanxi science and technology plan projects (2011K16-02-05), Xi’an key scientific and technological project (NC1116 (1)) and Innovation funds of graduate programs, SNNU (2013CXS017). The authors thank Dr. Leitch IJ (Royal Botanic Gardens, UK) for her kind revisions on the manuscript.
References
- Apg III . 2009. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot J Linn Soc. 161(S,2):105–121.
- Bai CK , Alverson WS , Follansbee A , Waller MD . 2012. New reports of nuclear DNA content for 407 vascular plant taxa from the United States. Ann Bot. 110(8):1623–1629.
- Bai CK , Wen MM , Zhang LJ , Li GS . 2013. Genetic diversity and sampling strategy of Scutellaria baicalensis germplasm resources based on ISSR. Genet Resour Crop Evol.60:1673–1685.
- Beaulieu JM , Leitch IJ , Knight CA . 2007. Genome size evolution in relation to leaf strategy and metabolic rates revisited. Ann Bot. 99(3):495–505.
- Beaulieu JM , Leitch IJ , Patel S , Pendharkar A , Knight CA . 2008. Genome size is a strong predictor of cell size and stomatal density in angiosperms. New Phytol. 179(4):975–986.
- Beaulieu JM , Moles AT , Leitch IJ , Bennett MD , Dickie JB , Knight CA . 2007. Correlated evolution of genome size and seed mass. New Phytol. 173(2):422–437.
- Beaulieu JM , Smith S , Leitch IJ . 2010. On the tempo of genome size evolution in angiosperms. J Bot. 2010:989152. http://dx.doi.org/10.1155/2010/989152
- Bennett W , Lubienski M , Korner Korner S , Steinberg M. 2005. Triploidy in Equisetum subgenus Hippochaete (Equisetaceae, Pteridophyta). Ann Bot. 95(5):807–815.
- Bennett MD , Bhandol P , Leitch IJ . 2000. Nuclear DNA amounts in angiosperms and their modern uses: 807 new estimates. Ann Bot. 86(4):859–909.
- Bennett MD , Leitch IJ . 1997. Nuclear DNA amounts in angiosperms: 583 new estimates. Ann Bot. 80(2):169–196.
- Bennett MD , Leitch IJ . 2005. Nuclear DNA amounts in angiosperms: progress, problems and prospects. Ann Bot. 95(1):45–90.
- Bennett MD , Leitch IJ . 2011. Nuclear DNA amounts in angiosperms: targets, trends and tomorrow. Ann Bot. 107(3):467–590.
- Bennett MD , Leitch IJ . 2012. Plant DNA C-values database [database]. http://data.kew.org/cvalues. Release 6.0. [updated 2012 December; cited 2013 March 30].
- Bennett MD , Smith JB . 1976. Nuclear DNA amounts in angiosperms. Philos T Roy Soc B. 274(933):227–274.
- Bharathan G , Lambert G , Galbraith DW . 1994. Nuclear DNA content of monocotyledons and related taxa. Am J Bot. 81(3):381–386.
- Chung KS , Hipp AL , Roalson EH . 2012. Chromosome number evolves independently of genome size in a clade with nonlocalized centromeres (Carex: Cyperaceae). Evolution. 66(9):2708–2722.
- Das AB , Mallick R . 1989. Variation in karyotype and nuclear DNA content in different varieties of Foeniculum vulgare Mill. Cytologia. 54(1):129–134.
- Doležel J , Bartoš J , Voglmayr H , Greilhuber J . 2003. Nuclear DNA content and genome size of trout and human. Cytom Part A. 51(2)A:127–128.
- Doležel J , Binarova P , Lucretti S . 1989. Analysis of nuclear DNA content in plant cells by flow cytometry. Biol Plantarum. 31(2):113–120.
- Doležel J , Gohde W . 1995. Sex determination in dioecious plants Melandrium album and M. rubrum using high-resolution flow cytometry. Cytometry. 19(2):103–106.
- Doležel J , Greilhuber J . 2010. Nuclear genome size: Are we getting closer? Cytom Part A. 77(7):635–642.
- Doležel J , Greilhuber J , Suda J . 2007a. Estimation of nuclear DNA content in plants using flow cytometry. Nat Protoc. 2(9):2233–2244.
- Doležel J , Greilhuber J , Suda J . 2007b. Flow cytometry with plants: an overview. In: Doležel J , Greilhuber J , Suda J , eds. Flow cytometry with plant cells. Weinheim: Wiley, 41–90.
- Fay MF , Cowan RS , Leitch IJ . 2005. The effects of nuclear DNA content (C-value) on the quality and utility of AFLP fingerprints. Ann Bot. 95(1):237–246.
- Franks PJ , Freckleton RP , Beaulieu JM , Leitch IJ , Beerling DJ . 2012. Megacycles of atmospheric carbon dioxide concentration correlate with fossil plant genome size. Philos T Roy Soc B. 367(1588):556–564.
- Greilhuber J , Borsch T , Müller K , Worberg A , Porembski S , Barthlott W . 2006. Smallest angiosperm genomes found in Lentibulariaceae with chromosomes of bacterial size. Plant Bio. 8(6):770–777.
- Greilhuber J , Leitch IJ . 2013. Genome size and the phenotype. In: Leitch IJ , Greilhuber J , Doležel J , Wendel JF , editors. Plant genome diversity. Vol. 2, Physical structure, behaviour and evolution of plant genomes. Vienna: Springer. p. 323–344.
- Greilhuber J , Temsch E , Loureiro J . 2007. Nuclear DNA content measurement. In: Doležel J , Greilhuber J , Suda J , eds. Flow cytometry with plant cells. Weinheim: Wiley, 67–101.
- Grime JP , Mowforth MA . 1982. Variation in genome size – an ecological interpretation. Nature. 299(5879):151–153.
- Hanson L , McMahon KA , Johnson MAT , Bennett MD . 2001. First nuclear DNA C-values for another 25 angiosperm families. Ann Bot. 88(5):851–858.
- Hodgson JG , Sharafi M , Jalili A , Díaz S , Montserrat-Martí G , Palmer C , Cerabolini B , Pierce S , Hamzehee B , Asri Y , et al . 2010. Stomatal vs. genome size in angiosperms: the somatic tail wagging the genomic dog? Ann Bot. 105(4):573–584.
- Houben A , Demidov D , Gernand D , Meister A , Leach CR , Schubert I . 2003. Methylation of histone H3 in euchromatin of plant chromosomes depends on basic nuclear DNA content. Plant J. 33(6):967–973.
- Janoušek B , Hobza R , Vyskot B . 2013. Chromosomes and sex differentiation. In: Leitch IJ , Greilhuber J , Doležel J , Wendel JF , editors. Plant genome diversity, Vol. 2. Physical structure, behavior, and evolution of plant genomes. Vienna : Springer. p. 167–186.
- Kelly LJ , Leitch IJ . 2011. Exploring giant plant genomes with next-generation sequencing technology. Chromosome Res. 19(7):939–953.
- Kelly LJ , Leitch AR , Fay MF , Renny-Byfield S , Pellicer J , Macas J , Leitch IJ . 2012. Why size really matters when sequencing plant genomes. Plant Ecol Divers. 5(4):415–425.
- Leitch AR , Leitch IJ . 2012. Ecological and genetic factors linked to contrasting genome dynamics in seed plants. New Phytol. 194(3):629–646.
- Leitch IJ , Bennett MD . 2007. Genome size and its uses: the impact of flow cytometry. In: Doležel J , Greilhuber J , Suda J , eds. Flow cytometry with plant cells. Weinheim: Wiley, 153–176.
- Leitch IJ , Bennett MD , Chase MW , Leitch AR , Fay MF . 2010. Genome size dynamics and evolution in monocots. J Bot. 2010:862516. http://dx.doi.org/10.1155/2010/862516.
- Leitch IJ , Kahandawala I , Suda J . 2009. Genome size diversity in orchids: consequences and evolution. Ann Bot. 104(3):469–481.
- Leitch IJ , Soltis DE , Soltis PS , Bennett MD . 2005. Evolution of DNA amounts across land plants (Embryophyta). Ann Bot. 95(1):207–217.
- Li GS , Zhang LJ , Bai CK . 2012. Chinese Cornus officinalis: genetic resources, genetic diversity and core collection. Genet Resour Crop Evol. 59(8):1659–1671.
- Loureiro J , Rodriguez E , Doležel J , Santos C . 2006. Comparison of four nuclear isolation buffers for plant DNA flow cytometry. Ann Bot. 98(3):679–689.
- Loureiro J , Rodriguez E , Doležel J , Santos C. 2007. Two new nuclear isolation buffers for plant DNA flow cytometry: a test with 37 species. Ann Bot. 100(4):875–888.
- Loureiro J , Suda J , Doležel J . 2007. FLOWer: A plant DNA flow cytometry database. In: Doležel J , Greilhuber J , Suda J , eds. Flow cytometry with plant cells. Weinheim: Wiley, 423–438.
- Nagl W , Habermann T , Fusenig HP . 1977. Nuclear DNA contents four primitive angiosperms. Plant Syst Evol. 127(2–3):103–105.
- Ohri D , Kumar A , Pal M . 1986. Correlations between 2C DNA values and habit in Cassia (Leguminosae: Caesalpinioideae). Plant Syst Evol. 153(3–4):223–227.
- Ormerod MG . 2008. Data analysis. In: Ormerod MG , editor. Flow cytometry: a basic introduction. Los Angeles: De Novo Software. http://flowbook.denovosoftware.com/Flow_Book/Chapter_4%3a_Data_Analysis
- Otto F . 1990. DAPI staining of fixed cells for high-resolution flow cytometry of nuclear DNA. In: Crissman HA , Darzynkiewicz Z , eds. Methods in cell biology. vol 33. New York, NY: Academic Press, 105–110.
- Pellicer J , Fay MF , Leitch IJ . 2010. The largest eukaryotic genome of them all? Bot Linn Soc. 164(1):10–15.
- Stevens PF [website]. 2012. Angiosperm phylogeny website, version 9. [updated 2008 June, with continual updates; cited 2013 March 30] www.mobot.org/MOBOT/research/APweb/
- Suda J , Leitch IJ . 2010. The quest for suitable reference standards in genome size research. Cytom Part A. 77(8)A:717–720.
- Swift H . 1950. The constancy of desoxyribose nucleic acid in plant nuclei. PNAS. 36(11):643–654.
- Temsch EM , Temsch W , Ehrendorfer-Schratt L , Greilhuber J. 2010. Heavy metal pollution, selection, and genome size: The species of the Žerjav study revisited with flow cytometry. J Bot 2010:596542, The Doi is “http://dx.doi.org/10.1155/2010/596542”
- Tian P . 2011. Where West meets East. Nature. 480(7378):S84–S86.
- Tu YY . 2011. The discovery of artemisinin (qinghaosu) and gifts from Chinese medicine. Nat Med. 17(10):1217–1220.
- Vallès J , Canela MÁ , Garcia S , Hidalgo O , Pellicer J , Jiménez IS , Siljak YS , Vitales GD . 2013. Genome size variation and evolution in the family Asteraceae. Caryologia. doi:10.1080/00087114.2013.829690.
- Vesely P , Bures P , Smarda P , Pavlicek T . 2011. Genome size and DNA base composition of geophytes: the mirror of phenology and ecology? Ann Bot. 109(1):65–75.
- Xu ZG . 2011. Modernization: One step at a time. Nature. 480(7378):S90–S92.
- Zonneveld BJM , Leitch IJ , Bennett MD . 2005. First nuclear DNA amounts in more than 300 angiosperms. Ann Bot. 96(2):229–244.
- Zonneveld BJM , Van Iren F . 2001. Genome size and pollen viability as taxonomic criteria: Application to the genus Hosta. Plant Bio 3(2):176–185.