Abstract
Ideograms are diagrammatic or idealized representations of chromosomes, showing their relative size, homologous groups and cytogenetic landmarks. Chromosomal analysis of cytological preparations involves calculation of karyotypic parameters and generation of ideograms. We generated IdeoKar, a free and user-friendly karyotype analysis software that facilitates data collection from different digital images of metaphase chromosome spreads and calculates a wide variety of chromosomal and karyotypic parameters along with the related standard errors which can be saved as a Microsoft Excel file. This software is also capable of drawing ideograms of both diploid and allopolyploid species in a single image file. IdeoKar is designed to fast and efficient karyotype analysis and ideogram generation. It can be used with personal computers without any specific hardware. This software and related documentation is freely available at the website of the University of Kurdistan at: http://agri.uok.ac.ir/ideokar/index.html.
Introduction
Each species has a characteristic chromosome complement, its karyotype, which represents the phenotypic appearance of the somatic chromosomes (Levitzky Citation1931). Ideograms are diagrammatic representations of the chromosomes showing their relative size, homologous groups and chromosomal landmarks. Ideograms are often used in many journal articles on a regular basis and in web interfaces to visualize genome and chromosomal information.
Karyotype analysis involves measurement of characteristics such as chromosome number, size and symmetry, as well as position and type of chromosomal landmarks such as centromeres, secondary constrictions, or repetitive DNA types (Heslop‐Harrison and Schwarzacher Citation2011). Such characteristics then may be used to distinguish homologous chromosomes. By comparing the chromosomes of different taxa much can be learned about patterns and mechanisms of karyotype evolution and its significance for diversification and speciation (Weiss-Schneeweiss et al. Citation2008).
Karyotype analyses have studied characteristics such as C- or N-banding patterns (Gill et al. Citation1991), in situ hybridization (ISH) patterns of repeated sequences (Mirzaghaderi et al. Citation2014), ISH of single copy sequences (Shen et al. Citation1987), or immunofluorescent localization of proteins on chromosomes (Anderson et al. Citation1999). Size and location of such landmarks are often quantifiable and are reflected in an ideogram.
Traditional measurement of metaphase spreads and collecting the data into tables is a slow and tedious process requiring intermediate photographic steps, manual statistical analysis and manual drawing of the ideogram image. Interest in applying automation to analysis of metaphase chromosomes dates back four decades and several computer applications have been developed to facilitate accurate karyotype analyses (Gilbert Citation1966; Green et al. Citation1980, Citation1984; James Citation1983; Fukui Citation1986; Armstrong et al. Citation1987; Oud et al. Citation1987; König and Ebert Citation1997; Reeves Citation2001). However, there is neither free software nor a web server to allow users to calculate chromosomal and karyotypic parameters and to build ideograms. Therefore, there is a need for a free and user-friendly software that can address these needs, for high quality karyotypic analysis. We developed IdeoKar software which allows users to facilitate chromosomal measurements and build ideograms from digitally captured or scanned images. The generated ideogram is a high-quality image that is suitable for poster presentation, projector screen presentation and journal publication.
Materials and methods
Overall software design and architecture
IdeoKar software is used for calculation of chromosomal and karyotypic parameters and to build ideograms. The software was written in C# programming language and consists of three main windows (Figure ). The core window is used for opening images, scale bar definition, chromosome tracing and numbering and chromosomal landmark definition. The second window is used for presentation of chromosomal and karyotypic parameters. The third window is used for showing the generated ideogram of the traced chromosomes (Figure ). Providing that IdeoKar is calibrated, a scale bar corresponding to 5 μm is placed on the ideogram.
Figure 1. (Color online) Using IdeoKar to trace different chromosomes of Nigella sativa L. Regions of the centromere in chromosomes are marked in red while regions of secondary constrictions are marked in blue.
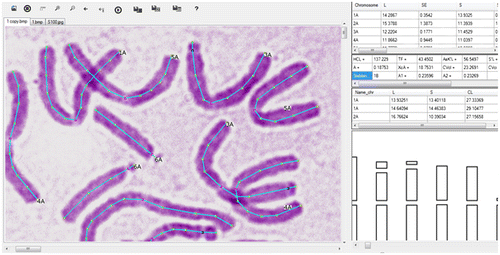
Figure 2. Ideogram of Nigella sativa, generated by IdeoKar after tracing two different metaphase images. Providing that IdeoKar is calibrated, a scale bar corresponding to 5 μm is placed on the ideogram.
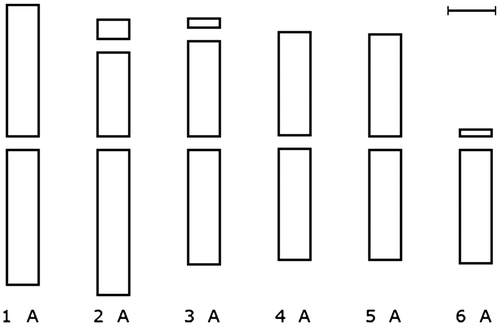
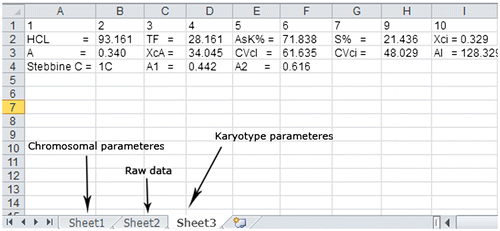
The parameters that are calculated are described in Table . After tracing chromosomes, all the related data in the tables can be stored in a Microsoft Excel file which includes three tabs. The nomenclature used for the description of chromosome type or morphology is that proposed by Levan et al. (Citation1964) in which the type of chromosome is defined based on mean of arm ratio (Table ). The chromosomal parameters are stored in the first tab, raw data are stored in the second tab and the karyotype parameters are stored in the third tab (Figure ).
Table 1. Measures and formulae of chromosomal parameters and karyotype asymmetry indices used in the IdeoKar program.
Table 2. Nomenclature of chromosomes based on Levan et al. Citation1964.
Table 3. Classification of karyotypes in relation to their degree of asymmetry according to Stebbins (Citation1971).
Figure 3. After tracing chromosomes, all the related data in the tables can be stored in an Excel file which includes three tabs. The chromosomal parameters are stored in the first tab, raw data are stored in the second tab and the karyotype parameters are stored in the third tab.
At present, MicroMeasure (Reeves Citation2001) is the only free computer program that is available for the collection and analysis of cytogenetic data. The IdeoKar software has the following advantages over MicroMeasure. First, there is the ability to calibrate the software based on the actual length of a known distance on the image so that all subsequent calculations can be in micrometers. The second advantage is the ability to calculate mean and standard deviation of chromosomal parameters of all chromosomes of the same homologous group. These homologous chromosomes may be traced in the same image and/or in the different opened images. After tracing chromosomes and pressing the Run key, different length calculations and chromosomal parameters will appear in the data sheet of the software along with their standard errors. The data can be stored in the form of an Excel file format. This data will be used to generate an ideogram image which can also be saved as any of the pixel based image file formats.
Software and hardware requirements
IdeoKar is designed for standard personal computers with the operating systems Microsoft Windows 7 or 8. There is no requirement to any specific type of hardware.
Length measurements and point marks
In order to trace chromosomes, the software asks for the number and homologous group of each chromosome in the metaphase spread. This requires that the researcher distinguishes the number of the homologous group and the genome that each particular chromosome belongs to. The user first opens different metaphase images belonging to the same or different species, ecotypes or individuals. All images are opened in separate tabs. Then in each tab the user traces chromosomes on the computer screen using mouse clicks and holds Ctrl + specific keys in specific points such as the centromere, band regions, secondary constrictions or 45S rDNA, 5S rDNA and chromosome ends to introduce them to the program. A small window is automatically opened after tracing each chromosome, requesting the number of the chromosome and its related genome. This chromosome tracing may be extended to any other opened image in the software.
Biological observations show that whole or arm lengths of two homologous chromosomes can be of very different sizes because they are affected by many factors such as the chromosome preparation method. So the result for arm size comparison can only support other results such as banding patterns rather than become a main factor. For this reason, some rules were considered when drawing ideograms including: (1). If there is no landmark or banding pattern in a homologous group, the short and long arms in that homologous group are selected based on length. (2). If there is a 5S rDNA locus, 45S rDNA locus or banding pattern in a homologous group, arms with a specific rDNA locus or banding pattern are considered as homologous arms even if the chromosomes have different arm ratios.
Sample analyses
To demonstrate how IdeoKar works, digital images of black cumin (Nigella sativa L.) were used (Figure ). The images were captured directly from a digital camera connected to a microscope. Chromosome tracking was conducted with IdeoKar software. Some chromosomal features such as centromeres and secondary constrictions. were generally treated as discrete points that occur along the chromosomes.
Figure shows an ideogram image which was generated by IdeoKar by tracing the metaphase chromosomes of Nigella sativa L.
Results and discussion
IdeoKar is a powerful and extensible tool for the collection of cytogenetic data. This software was designed to reduce waiting and wasted time and to be user friendly. Users have the opportunity to draw ideograms and obtain chromosomal parameters from the images of their particular metaphase spreads without the need to perform time consuming calculations. IdeoKar supports bmp, gif, ico, jpg, mng, svg, tga, and tif formats so there should be no need to convert the originally acquired image.
IdeoKar has a simple environment and it is capable of simultaneously drawing different ideograms of different species, subspecies or individuals in a single image file. This capability enables the user to generate ideograms of allopolyploid plants as well. Allopolyploids have two or more different genomes in the cell and are very common among plant species. Chromosomes of each genome in the ideogram of such an allopolyploid are shown in separate rows so, there would be more than one row of chromosomes in the generated ideogram.
Integration with Microsoft Excel further extends the users’ options, providing for further quantitative and statistical analysis either within Excel or with any of the numerous computer applications compatible with Excel. IdeoKar’s compatibility with standard personal computers eliminates some of the need for more costly and more complicated software and equipment. IdeoKar does not have an option to automatically distinguish homologous chromosomes. Thus the user needs to distinguish the number of homologous group and the genome that each particular chromosome belongs to. This software and related documentation is freely available on the website of the University of Kurdistan at: http://agri.uok.ac.ir/ideokar/index.html.
References
- Anderson LK, Reeves A, Webb LM, Ashley T. 1999. Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics. 151(4):1569–1579.
- Arano H. 1963. Cytological studies in subfamily Carduoideae (Compositae) of Japan. IX. Bot Mag. 76: 32.
- Armstrong K, Craig IL, Merritt C. 1987. Hordeum chilense (2n = 14) computer-assisted Giemsa karyotypes. Genome. 29(5):683–688.
- Fukui K. 1986. Standardization of karyotyping plant chromosomes by a newly developed chromosome image analyzing system (CHIAS). Theor Appl Genet. 72(1): 27–32.
- Gill B, Friebe B, Endo T. 1991. Standard karyotype and nomenclature system for description of chromosome bands and structural aberrations in wheat (Triticum aestivum). Genome. 34(5): 830–839.
- Gilbert CW. 1966. A computer programme for the analysis of human chromosomes. Nature. 212(5069):1437–1440.
- Green DM, Bogart JP, Anthony E, Genner D. 1980. An interactive, microcomputer-based karyotype analysis system for phylogenetic cytotaxonomy. Comput Biol Med. 10(4): 219–227.
- Green DM, Myers PZ, Reyna DL. 1984. CHROMPAC III: an improved package for microcomputer assisted analysis of karyotypes. J Hered. 75(2): 143–143.
- Heslop-Harrison J, Schwarzacher T. 2011. Organisation of the plant genome in chromosomes. Plant J. 66(1):18–33.
- Huziwara Y. 1962. Karyotype analysis in some genera of Compositae. VIII. Further studies on the chromosomes of Aster. Am J of Bot. 49(2):116–119.
- James M. 1983. A BASIC computer program for chromosome measurement and analysis. J Hered. 74(4): 304.
- König C, Ebert I. 1997. Computer-aided quantitative analysis of banded karyotypes, exemplified in C-banded Hyacinthoides italica sl (Hyacinthaceae). Caryologia. 50(2): 105–116.
- Levan A, Fredga K, Sandberg AA. 1964. Nomenclature for centromeric position on chromosomes. Hereditas. 52(2):201–220.
- Levitzky GA. 1931. The karyotype in systematics. Bull Appl Bot, Genet Plant Breed. 27: 220–240.
- Mirzaghaderi G, Houben A, Badaeva E. 2014. Molecular-cytogenetic analysis of Aegilops triuncialis and identification of its chromosomes in the background of wheat. Mol Cytogenet. 7:91.
- Oud J, Kakes P, De Jong J. 1987. Computerized analysis of chromosomal parameters in karyotype studies. Theor Appl Genet. 73(5): 630–634.
- Paszko B. 2006. A critical review and a new proposal of karyotype asymmetry indices. Plant Syst Evol. 258(1): 39–48.
- Reeves A. 2001. MicroMeasure: a new computer program for the collection and analysis of cytogenetic data. Genome. 44(3): 439–443.
- Romero-Zarco C. 1986. A new method for estimating karyotype asymmetry. Taxon. 35(3):526–530.
- Shen D-L, Wang Z-F, Wu M. 1987. Gene mapping on maize pachytene hormosomes by in situ hybridization. Chromosoma. 95(5):311–314.
- Stebbins GL. 1971. Chromosomal evolution in higher plants, London: Edward Arnold.
- Watanabe K, Yahara T, Denda T, Kosuge K. 1999. Chromosomal evolution in the genus Brachyscome (Asteraceae, Astereae): statistical tests regarding correlation between changes in karyotype and habit using phylogenetic information. J Plant Res. 112(2): 145–161.
- Weiss-Schneeweiss H, Tremetsberger K, Schneeweiss GM, Parker JS, Stuessy TF. 2008. Karyotype diversification and evolution in diploid and polyploid South American Hypochaeris (Asteraceae) inferred from rDNA localization and genetic fingerprint data. Ann bot. 101(7): 909–918.
